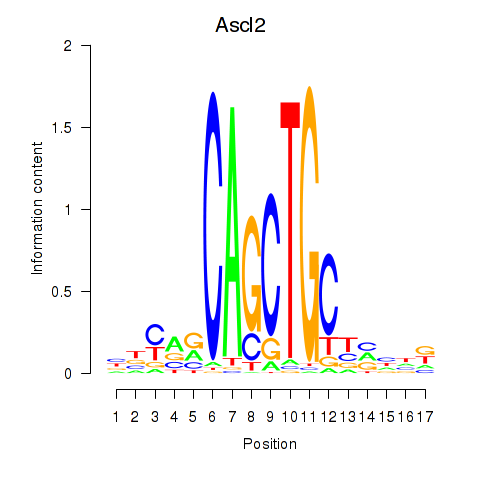
| 1.9 |
5.6 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.8 |
2.4 |
GO:0072076 |
hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.8 |
3.2 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.7 |
2.2 |
GO:2000097 |
regulation of smooth muscle cell-matrix adhesion(GO:2000097) negative regulation of lymphocyte migration(GO:2000402) |
| 0.7 |
2.2 |
GO:0046544 |
development of secondary male sexual characteristics(GO:0046544) |
| 0.7 |
2.9 |
GO:0010957 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.7 |
0.7 |
GO:0048389 |
intermediate mesoderm development(GO:0048389) pattern specification involved in kidney development(GO:0061004) pattern specification involved in mesonephros development(GO:0061227) renal system pattern specification(GO:0072048) anterior/posterior pattern specification involved in kidney development(GO:0072098) ureter urothelium development(GO:0072190) |
| 0.7 |
2.8 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.7 |
2.0 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.6 |
1.9 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.6 |
2.5 |
GO:0051599 |
response to hydrostatic pressure(GO:0051599) |
| 0.6 |
1.2 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.6 |
1.8 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.6 |
1.7 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.6 |
4.5 |
GO:1903689 |
regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.5 |
3.6 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.5 |
1.5 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.4 |
1.3 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.4 |
2.7 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.4 |
3.0 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.4 |
2.0 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.4 |
5.5 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.4 |
1.2 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.4 |
2.1 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.4 |
1.1 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.3 |
1.0 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.3 |
1.0 |
GO:0002586 |
regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.3 |
2.9 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 |
0.7 |
GO:0072197 |
ureter morphogenesis(GO:0072197) |
| 0.3 |
1.3 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 |
2.2 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.3 |
1.2 |
GO:0010046 |
arginine biosynthetic process(GO:0006526) response to mycotoxin(GO:0010046) |
| 0.3 |
1.4 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 |
0.8 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.3 |
1.1 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.3 |
0.8 |
GO:0046618 |
drug export(GO:0046618) |
| 0.3 |
0.8 |
GO:0046959 |
habituation(GO:0046959) |
| 0.3 |
1.0 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 |
2.5 |
GO:1901550 |
regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.2 |
1.2 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.2 |
0.7 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 |
1.9 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.2 |
1.0 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 |
0.7 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.2 |
1.4 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 |
2.4 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 |
0.7 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.2 |
2.6 |
GO:0001977 |
renal system process involved in regulation of blood volume(GO:0001977) |
| 0.2 |
0.2 |
GO:0090080 |
positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 |
1.2 |
GO:0006447 |
regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 |
0.4 |
GO:0042359 |
vitamin D metabolic process(GO:0042359) |
| 0.2 |
2.6 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 |
0.6 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.2 |
0.6 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.2 |
1.5 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 |
2.4 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 |
0.5 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 |
5.6 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.2 |
0.9 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 |
0.5 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) |
| 0.2 |
3.2 |
GO:0048368 |
lateral mesoderm development(GO:0048368) |
| 0.2 |
0.8 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.2 |
0.7 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 |
0.7 |
GO:1900864 |
mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 |
2.3 |
GO:0090205 |
positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.2 |
0.8 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 |
0.6 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.2 |
0.6 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 |
0.6 |
GO:0051305 |
chromosome movement towards spindle pole(GO:0051305) |
| 0.2 |
5.7 |
GO:0010862 |
positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.2 |
0.5 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 |
1.1 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
0.9 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.4 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 |
0.5 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.1 |
1.9 |
GO:0090308 |
regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 |
0.4 |
GO:1900107 |
regulation of nodal signaling pathway(GO:1900107) |
| 0.1 |
0.9 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.1 |
1.8 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
1.6 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.1 |
1.3 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.1 |
0.2 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.1 |
0.9 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.5 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
1.6 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 |
0.8 |
GO:0006573 |
valine metabolic process(GO:0006573) |
| 0.1 |
0.3 |
GO:0040009 |
nucleolus to nucleoplasm transport(GO:0032066) regulation of growth rate(GO:0040009) |
| 0.1 |
1.6 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
0.3 |
GO:0097278 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) complement-dependent cytotoxicity(GO:0097278) |
| 0.1 |
0.8 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 |
2.6 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 |
1.0 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.5 |
GO:0016560 |
protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 |
3.6 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.7 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
1.1 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.7 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
0.5 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 |
0.5 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.1 |
3.0 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
1.0 |
GO:0060586 |
multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.1 |
0.8 |
GO:0021678 |
third ventricle development(GO:0021678) |
| 0.1 |
0.6 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 |
0.3 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 |
0.7 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
1.4 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 |
0.7 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
0.4 |
GO:0040031 |
snRNA modification(GO:0040031) |
| 0.1 |
0.4 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.1 |
1.1 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.7 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 |
0.7 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
0.4 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.4 |
GO:0055091 |
phospholipid homeostasis(GO:0055091) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 |
0.5 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.1 |
0.2 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 |
0.2 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 |
0.1 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 0.1 |
1.0 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 |
0.5 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
1.4 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.5 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.8 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.1 |
GO:0072070 |
loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.0 |
0.3 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 |
0.1 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 |
1.0 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.6 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.4 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.1 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 |
0.7 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.5 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.6 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) |
| 0.0 |
0.3 |
GO:0046500 |
S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 |
0.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.1 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.0 |
1.3 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.8 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.9 |
GO:0071320 |
cellular response to cAMP(GO:0071320) |
| 0.0 |
1.5 |
GO:0030835 |
negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 |
0.2 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.7 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.5 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.6 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.8 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.0 |
0.3 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.5 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
2.8 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0061535 |
glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 |
0.3 |
GO:0033292 |
T-tubule organization(GO:0033292) |
| 0.0 |
2.4 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
0.0 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 |
0.7 |
GO:0009615 |
response to virus(GO:0009615) |
| 0.0 |
0.3 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
1.1 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 |
2.2 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
1.3 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 |
0.8 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.9 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
0.9 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
0.9 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.5 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.6 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
0.8 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 |
0.2 |
GO:0002031 |
G-protein coupled receptor internalization(GO:0002031) |
| 0.0 |
0.1 |
GO:0034134 |
toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 |
0.4 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 |
1.3 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
0.0 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 |
0.6 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.5 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
1.2 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.8 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.6 |
GO:0006805 |
xenobiotic metabolic process(GO:0006805) |
| 0.0 |
0.6 |
GO:0044380 |
protein localization to cytoskeleton(GO:0044380) |
| 0.0 |
0.4 |
GO:0006739 |
NADP metabolic process(GO:0006739) |
| 0.0 |
0.2 |
GO:0042921 |
glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 |
0.6 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.0 |
0.0 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 |
0.8 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 |
0.3 |
GO:0001709 |
cell fate determination(GO:0001709) |
| 0.0 |
0.4 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.8 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.0 |
0.6 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.7 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.4 |
GO:0016525 |
negative regulation of angiogenesis(GO:0016525) |
| 0.0 |
1.3 |
GO:0010921 |
regulation of phosphatase activity(GO:0010921) |
| 0.0 |
0.3 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 |
0.5 |
GO:0006893 |
Golgi to plasma membrane transport(GO:0006893) |
| 0.0 |
1.4 |
GO:0007160 |
cell-matrix adhesion(GO:0007160) |
| 0.0 |
0.0 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.0 |
0.1 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.0 |
0.7 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.2 |
GO:0044381 |
glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 |
0.1 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 |
1.3 |
GO:0019827 |
stem cell population maintenance(GO:0019827) |
| 0.0 |
0.5 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |