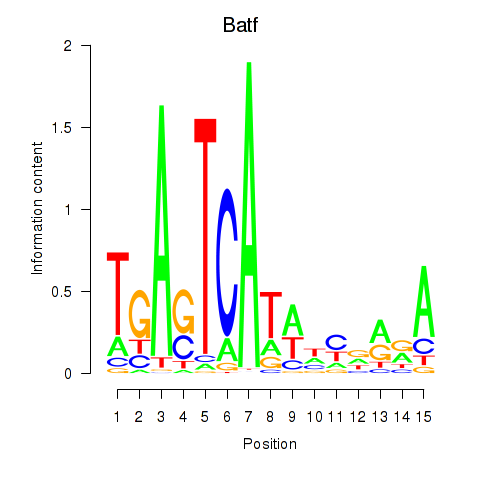
| 0.2 |
0.9 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.4 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.1 |
0.7 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.8 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 |
0.5 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
0.2 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 |
0.3 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
0.5 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
0.5 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.3 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.2 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.1 |
0.3 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
2.3 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.2 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 |
0.3 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.2 |
GO:0071397 |
cellular response to cholesterol(GO:0071397) |
| 0.0 |
0.0 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 |
0.4 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
0.1 |
GO:0009098 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 |
0.4 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
0.0 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.0 |
0.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |