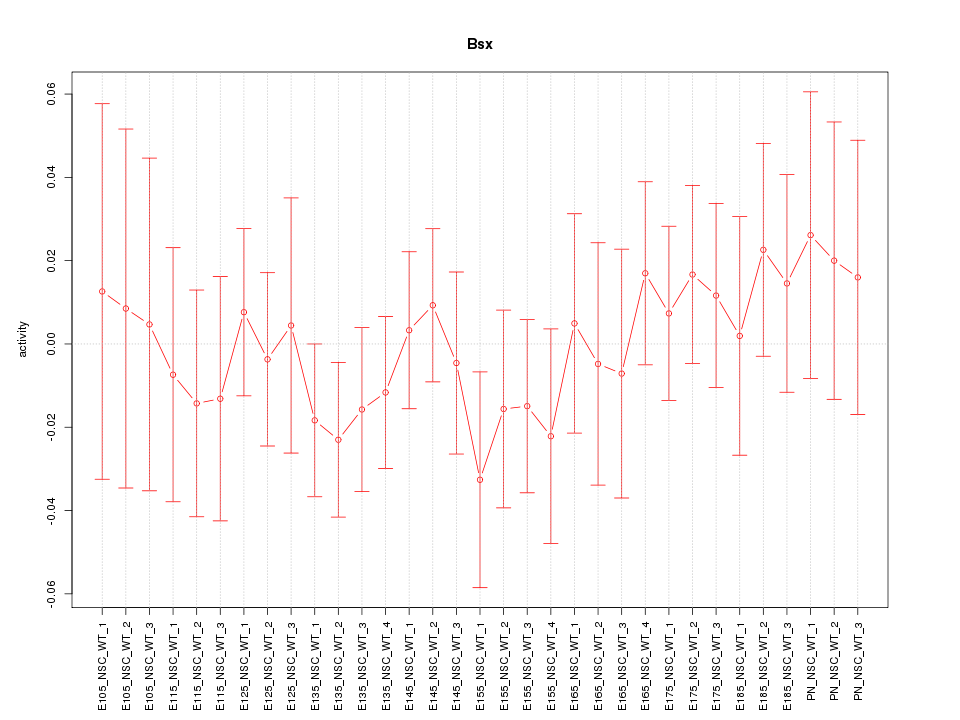
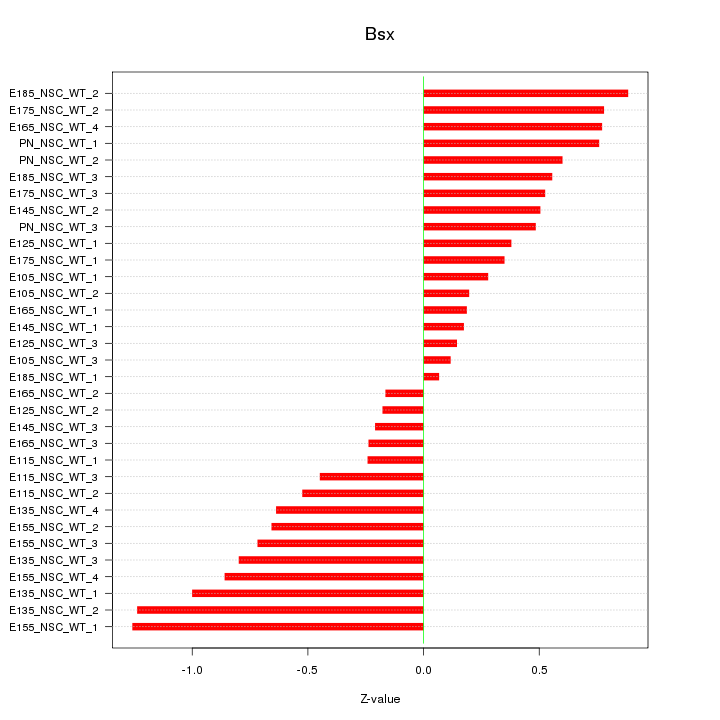
Motif ID: Bsx
Z-value: 0.604
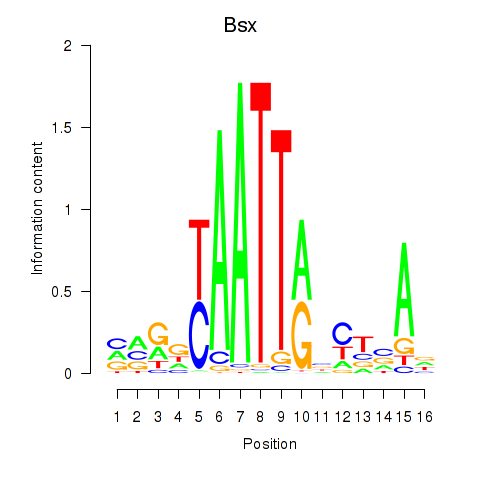
Transcription factors associated with Bsx:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bsx | ENSMUSG00000054360.3 | Bsx |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bsx | mm10_v2_chr9_+_40873981_40874127 | -0.27 | 1.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.3 | 1.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.3 | 0.9 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 0.9 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.3 | 1.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 2.4 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.2 | 1.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 1.6 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.9 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.4 | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.1 | 0.5 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.8 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 1.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.7 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 1.6 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 1.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 1.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0035633 | negative regulation of norepinephrine secretion(GO:0010700) maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.6 | GO:0043205 | fibril(GO:0043205) |
| 0.2 | 1.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 2.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.6 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 1.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.3 | 0.9 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 2.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.6 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 2.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |