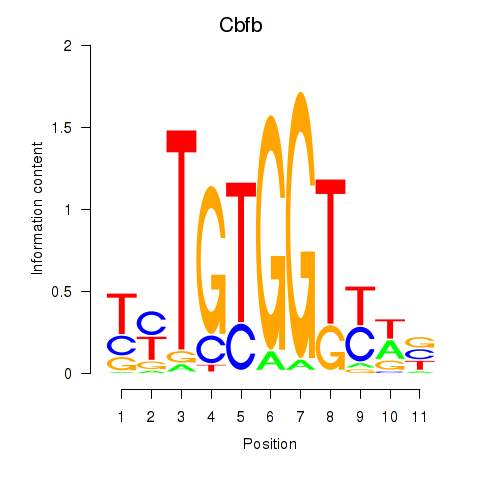
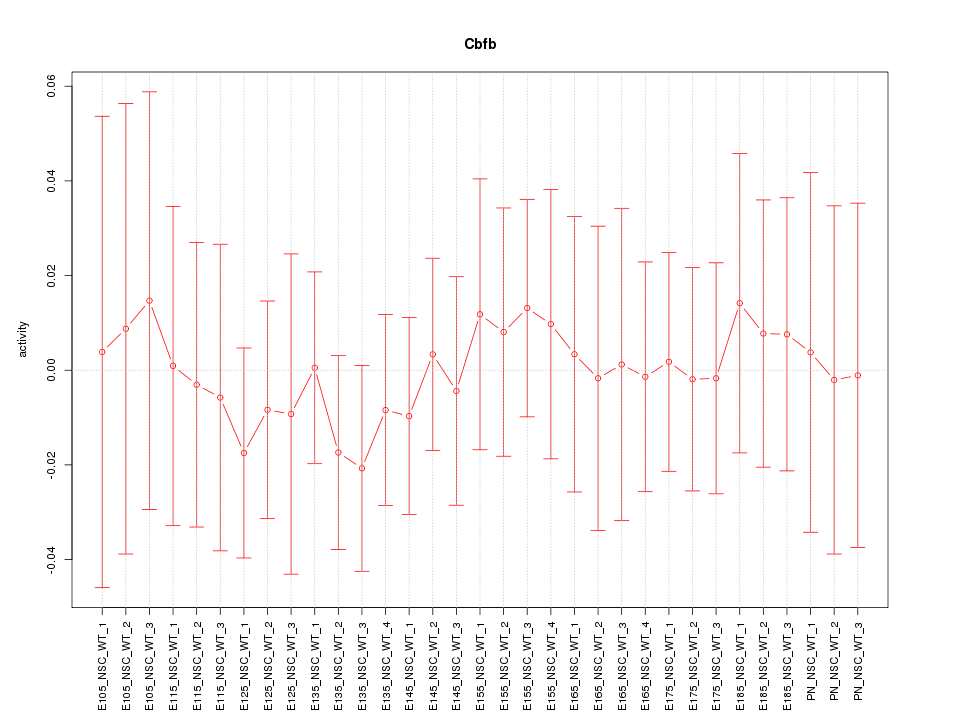
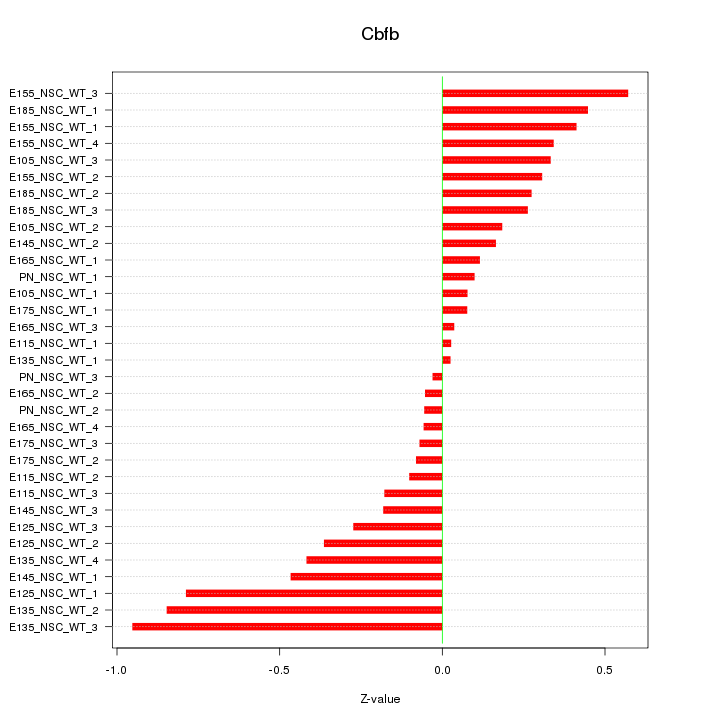
| 0.1 |
0.3 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 |
0.2 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) positive regulation of receptor binding(GO:1900122) |
| 0.0 |
0.2 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 |
0.1 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) |
| 0.0 |
0.2 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 |
0.1 |
GO:0036508 |
protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 |
0.1 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.0 |
0.1 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) regulation of microtubule motor activity(GO:2000574) |
| 0.0 |
0.4 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |