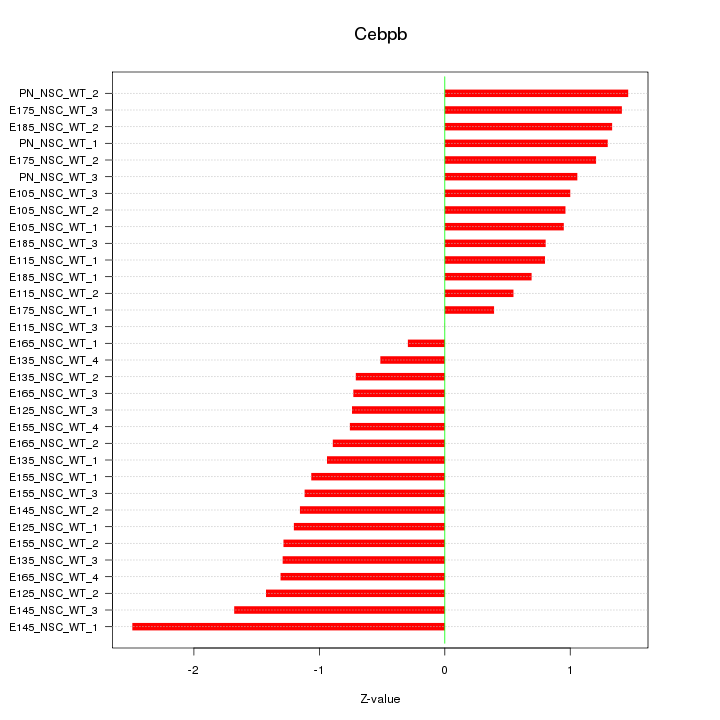
Motif ID: Cebpb
Z-value: 1.110
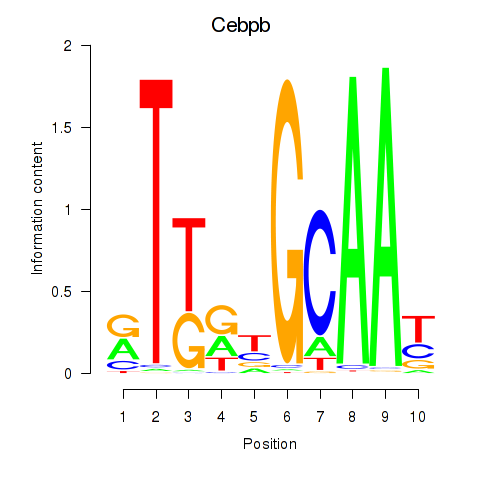
Transcription factors associated with Cebpb:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cebpb | ENSMUSG00000056501.3 | Cebpb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpb | mm10_v2_chr2_+_167688915_167688973 | 0.31 | 8.0e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 17.0 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 2.5 | 7.5 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 2.2 | 4.5 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 1.7 | 15.5 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 1.3 | 3.9 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.8 | 2.5 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.8 | 2.5 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.8 | 3.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.8 | 2.4 | GO:0008228 | opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.8 | 2.4 | GO:0045349 | regulation of dendritic cell cytokine production(GO:0002730) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.7 | 2.9 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.6 | 3.8 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.5 | 2.7 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.5 | 2.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 | 2.2 | GO:0015867 | ATP transport(GO:0015867) |
| 0.4 | 1.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.4 | 1.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.4 | 1.6 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 7.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.4 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 1.1 | GO:0010752 | negative regulation of antigen processing and presentation(GO:0002578) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) negative regulation of plasminogen activation(GO:0010757) |
| 0.4 | 1.8 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.4 | 1.4 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 | 3.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 1.3 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.3 | 1.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 | 1.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 2.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.3 | 2.9 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.3 | 0.8 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.3 | 1.8 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 2.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 1.7 | GO:0032782 | urea transport(GO:0015840) bile acid secretion(GO:0032782) urea transmembrane transport(GO:0071918) |
| 0.2 | 0.7 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.2 | 0.7 | GO:0010986 | complement receptor mediated signaling pathway(GO:0002430) GPI anchor release(GO:0006507) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 0.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 0.7 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 0.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 2.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.2 | 1.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 1.2 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.2 | 3.9 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.2 | 3.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 0.6 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 1.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.2 | 2.2 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 0.5 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 2.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 | 0.3 | GO:1900426 | positive regulation of defense response to bacterium(GO:1900426) |
| 0.2 | 0.2 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 | 0.9 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.7 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.1 | 1.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.0 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 8.6 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.1 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.8 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 1.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 1.4 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 5.0 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 1.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 1.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) megakaryocyte development(GO:0035855) |
| 0.1 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 1.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.1 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.4 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.7 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0007143 | female meiotic division(GO:0007143) rRNA transcription(GO:0009303) |
| 0.0 | 1.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.4 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.3 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 17.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 1.1 | 15.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.4 | 1.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 2.1 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 0.7 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 3.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.9 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 7.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 3.0 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 2.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 7.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 4.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 1.2 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 6.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 6.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.9 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 1.1 | 4.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 1.0 | 3.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.8 | 2.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.7 | 17.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.7 | 2.9 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.7 | 2.7 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.6 | 3.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 7.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.5 | 2.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.5 | 2.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 2.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 1.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.4 | 1.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.4 | 17.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 1.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 1.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 1.0 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.3 | 2.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 3.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.3 | 1.7 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.3 | 1.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.3 | 8.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.8 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.2 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 3.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 0.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 1.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.2 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 2.5 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.2 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 2.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 3.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 1.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 2.5 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 1.0 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 3.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.7 | GO:0015932 | nucleobase-containing compound transmembrane transporter activity(GO:0015932) |
| 0.0 | 1.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0017161 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |