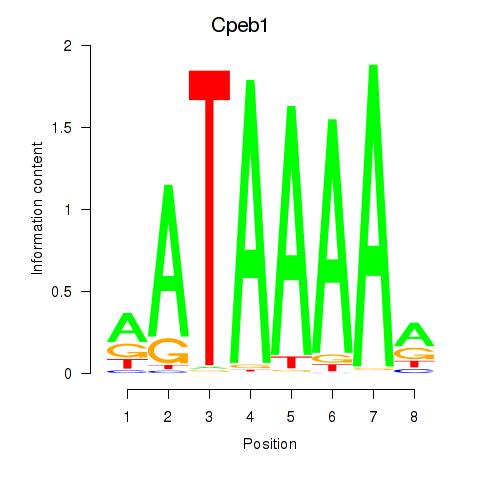
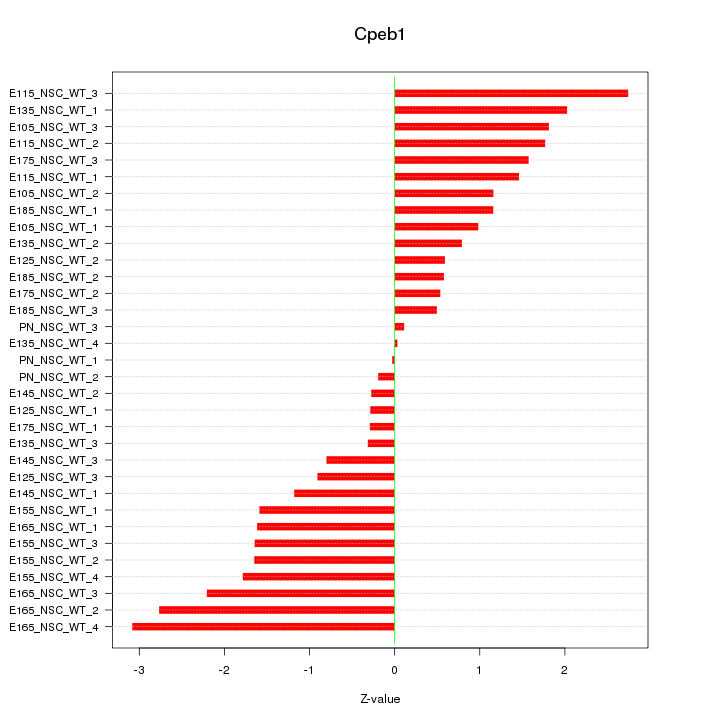
| 6.4 |
25.8 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 5.3 |
21.1 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 2.8 |
14.2 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 2.3 |
18.3 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 1.9 |
5.8 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.8 |
9.2 |
GO:0072015 |
ciliary body morphogenesis(GO:0061073) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 1.8 |
5.4 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.7 |
5.1 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 1.2 |
5.0 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.2 |
3.5 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 1.1 |
9.5 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 1.1 |
13.7 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.9 |
1.9 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.9 |
2.6 |
GO:0008582 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.8 |
2.4 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.8 |
7.9 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.8 |
5.5 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.7 |
2.1 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.7 |
6.4 |
GO:1902166 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.7 |
3.4 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.7 |
4.0 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.6 |
1.9 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.6 |
2.5 |
GO:0035878 |
nail development(GO:0035878) |
| 0.6 |
4.2 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.6 |
2.3 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.6 |
1.7 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.6 |
1.7 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.5 |
4.8 |
GO:0031642 |
negative regulation of myelination(GO:0031642) negative regulation of receptor catabolic process(GO:2000645) |
| 0.5 |
2.1 |
GO:1903898 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.5 |
2.6 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.5 |
3.6 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.5 |
2.5 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.5 |
5.0 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.5 |
2.3 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 |
1.8 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.4 |
2.1 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.4 |
1.2 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.4 |
2.7 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.4 |
5.1 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.4 |
1.4 |
GO:0045091 |
regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.3 |
1.0 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.3 |
1.0 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.3 |
1.0 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.3 |
1.6 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 |
1.6 |
GO:1900147 |
Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.3 |
2.9 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.3 |
2.2 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.3 |
0.6 |
GO:0046533 |
regulation of photoreceptor cell differentiation(GO:0046532) negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.3 |
0.9 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.3 |
4.5 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 |
2.0 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 |
0.6 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.3 |
1.4 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 |
3.0 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.3 |
0.5 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.3 |
1.3 |
GO:0060690 |
epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.3 |
1.8 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.3 |
1.3 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 |
9.7 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 |
1.0 |
GO:0006529 |
asparagine biosynthetic process(GO:0006529) |
| 0.2 |
4.5 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.2 |
1.9 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 |
0.2 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 |
1.1 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 |
0.7 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.2 |
2.8 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 |
2.6 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.2 |
2.3 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.2 |
0.4 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.2 |
0.6 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 |
1.0 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.2 |
0.8 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.2 |
1.0 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.2 |
1.0 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.2 |
1.0 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.2 |
2.3 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.2 |
1.5 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.2 |
3.3 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.2 |
0.6 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 |
2.2 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 |
0.9 |
GO:0035989 |
tendon development(GO:0035989) |
| 0.2 |
1.7 |
GO:0019511 |
peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 |
3.2 |
GO:0034142 |
toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.2 |
0.3 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.2 |
0.5 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.2 |
2.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.2 |
1.7 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 |
0.4 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 |
5.2 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 |
3.3 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
1.6 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
1.4 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 |
1.0 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
1.1 |
GO:0043518 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 |
1.1 |
GO:0097531 |
mast cell migration(GO:0097531) |
| 0.1 |
1.1 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.1 |
0.4 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
3.2 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 |
0.5 |
GO:2000427 |
regulation of apoptotic cell clearance(GO:2000425) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
0.7 |
GO:2001053 |
regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 |
1.0 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.1 |
1.7 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.1 |
0.3 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 |
6.6 |
GO:0051384 |
response to glucocorticoid(GO:0051384) |
| 0.1 |
2.9 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 |
1.1 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 |
2.5 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
5.8 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 |
0.4 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.1 |
2.0 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 |
0.2 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.1 |
0.8 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
1.6 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 |
3.2 |
GO:0051642 |
centrosome localization(GO:0051642) |
| 0.1 |
1.5 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.3 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
9.2 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.1 |
0.5 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 |
1.0 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 |
2.4 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.1 |
0.3 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.1 |
1.1 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.1 |
1.3 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 |
0.3 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 |
0.7 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 |
0.3 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
2.3 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.4 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
0.2 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 |
0.5 |
GO:0033572 |
transferrin transport(GO:0033572) |
| 0.1 |
2.4 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.1 |
0.2 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
0.3 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
2.4 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 |
0.2 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.0 |
5.2 |
GO:0070374 |
positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 |
4.2 |
GO:0048015 |
phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 |
0.4 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
1.1 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.0 |
1.4 |
GO:0007099 |
centriole replication(GO:0007099) |
| 0.0 |
1.9 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
1.2 |
GO:0060706 |
cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 |
1.0 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
1.6 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
1.1 |
GO:0006303 |
double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 |
0.1 |
GO:0097401 |
synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 |
0.3 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.5 |
GO:0006744 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
2.2 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.7 |
GO:0048599 |
oocyte development(GO:0048599) |
| 0.0 |
0.4 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.5 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
1.5 |
GO:0043627 |
response to estrogen(GO:0043627) |
| 0.0 |
0.3 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.5 |
GO:0044342 |
type B pancreatic cell proliferation(GO:0044342) |
| 0.0 |
0.9 |
GO:0021680 |
cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 |
1.2 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.1 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
1.2 |
GO:1902653 |
cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 |
0.4 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
1.3 |
GO:0050728 |
negative regulation of inflammatory response(GO:0050728) |
| 0.0 |
0.4 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.2 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.5 |
GO:0001947 |
heart looping(GO:0001947) |
| 0.0 |
1.4 |
GO:0046034 |
ATP metabolic process(GO:0046034) |
| 0.0 |
1.1 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.9 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.3 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
0.1 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.1 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
1.0 |
GO:0048593 |
camera-type eye morphogenesis(GO:0048593) |
| 0.0 |
0.0 |
GO:0045358 |
negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.0 |
1.6 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.1 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.3 |
GO:1901799 |
negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) |