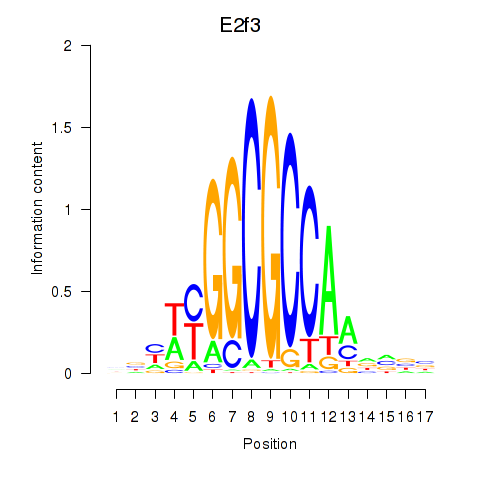
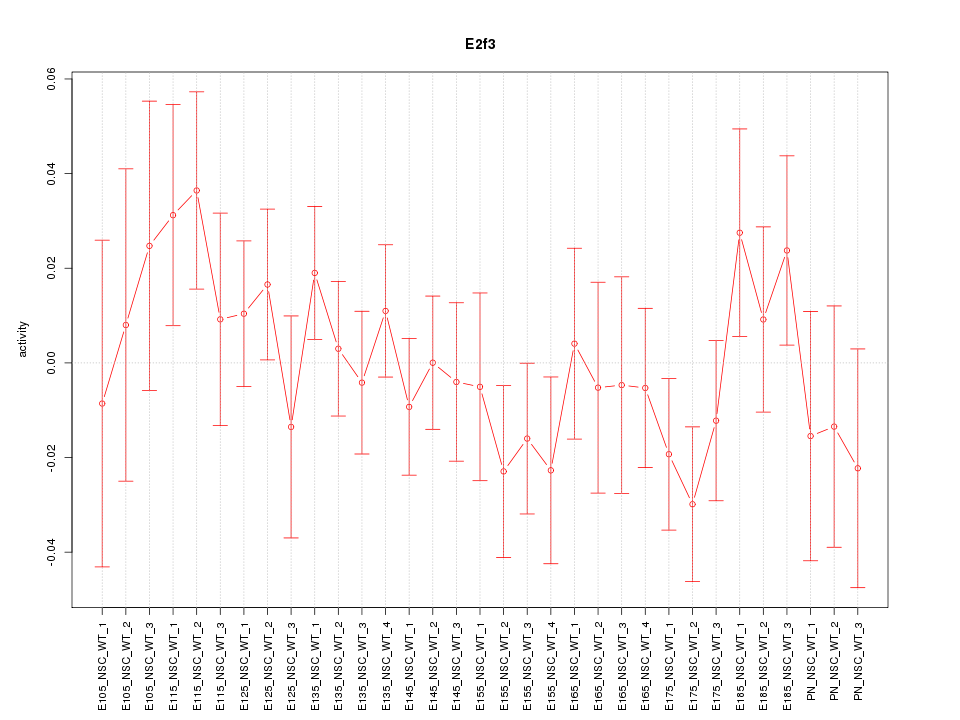
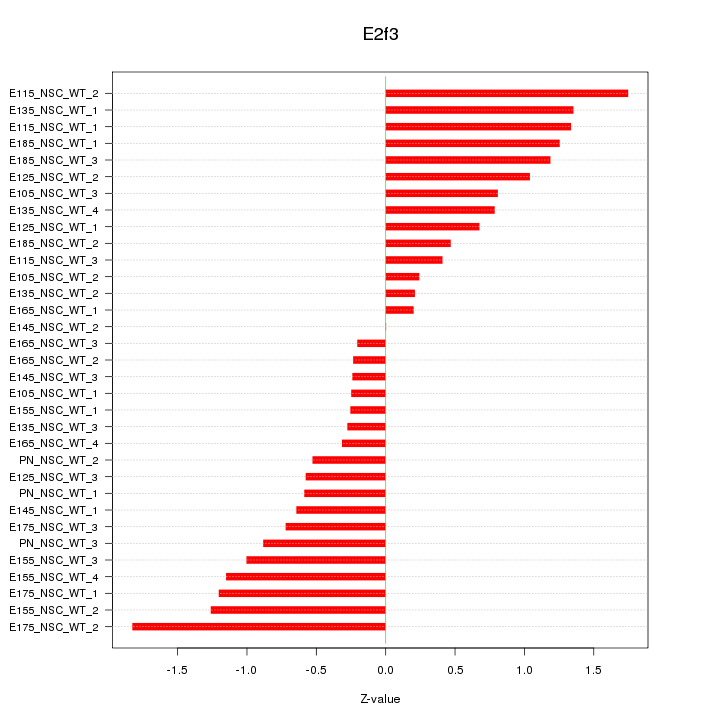
| 0.6 |
1.9 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.4 |
1.7 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.4 |
1.1 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.4 |
1.8 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.3 |
1.0 |
GO:0010752 |
negative regulation of antigen processing and presentation(GO:0002578) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) negative regulation of plasminogen activation(GO:0010757) |
| 0.2 |
1.0 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.2 |
2.4 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.2 |
1.0 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 |
0.2 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 |
0.7 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.2 |
0.7 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 |
0.7 |
GO:0000087 |
mitotic M phase(GO:0000087) mitotic cell cycle phase(GO:0098763) |
| 0.2 |
0.7 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 |
0.7 |
GO:0031990 |
mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.2 |
0.9 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 |
0.7 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 |
1.1 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 |
0.7 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.2 |
0.8 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.2 |
0.5 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 |
0.5 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.2 |
0.6 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.2 |
0.8 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 |
1.0 |
GO:0048478 |
replication fork protection(GO:0048478) |
| 0.1 |
4.0 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 |
0.8 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
1.3 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.1 |
4.5 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 |
0.6 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.1 |
1.0 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
1.0 |
GO:1901970 |
positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.1 |
0.9 |
GO:0033504 |
floor plate development(GO:0033504) |
| 0.1 |
0.3 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.5 |
GO:0046654 |
'de novo' IMP biosynthetic process(GO:0006189) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 |
0.4 |
GO:0035624 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) response to high density lipoprotein particle(GO:0055099) |
| 0.1 |
0.4 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.1 |
0.9 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 |
0.3 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.1 |
0.6 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 |
0.5 |
GO:1900016 |
negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.1 |
0.5 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 |
0.2 |
GO:0000965 |
mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.1 |
0.6 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.1 |
1.1 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 |
0.2 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 |
1.4 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.5 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 |
0.8 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.1 |
0.3 |
GO:0010387 |
COP9 signalosome assembly(GO:0010387) |
| 0.1 |
0.9 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
0.7 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
0.5 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.4 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
1.5 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
1.1 |
GO:0010574 |
regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.0 |
0.7 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.7 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.3 |
GO:0033262 |
regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 |
0.4 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.7 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
1.0 |
GO:0048246 |
macrophage chemotaxis(GO:0048246) |
| 0.0 |
0.4 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.9 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.2 |
GO:0042148 |
strand invasion(GO:0042148) |
| 0.0 |
0.9 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.0 |
0.5 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.4 |
GO:1904153 |
tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.5 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.1 |
GO:1900272 |
negative regulation of long-term synaptic potentiation(GO:1900272) regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 |
0.1 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 |
0.5 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.4 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
0.5 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:2000564 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 |
0.4 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.1 |
GO:0006296 |
nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 |
0.9 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.1 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.1 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 |
0.2 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.7 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.6 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
0.4 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.2 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.1 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 |
0.2 |
GO:2000773 |
negative regulation of cellular senescence(GO:2000773) |
| 0.0 |
0.2 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.3 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.4 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.7 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 |
0.1 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.6 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.3 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.1 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 |
0.5 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.5 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.7 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 |
0.2 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.3 |
GO:0071800 |
podosome assembly(GO:0071800) |
| 0.0 |
0.1 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.0 |
0.9 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.1 |
GO:1902534 |
single-organism membrane invagination(GO:1902534) |
| 0.0 |
0.5 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 |
0.3 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.1 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 |
0.4 |
GO:0043631 |
mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |