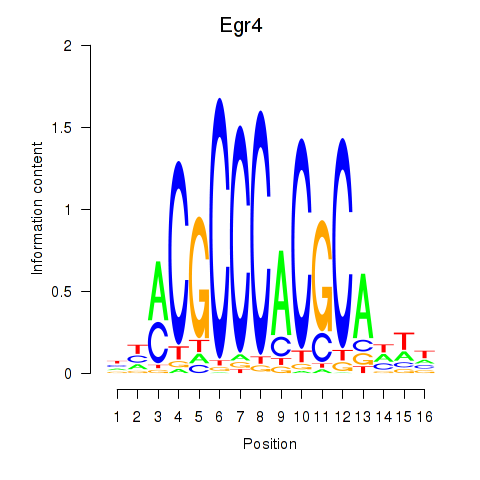
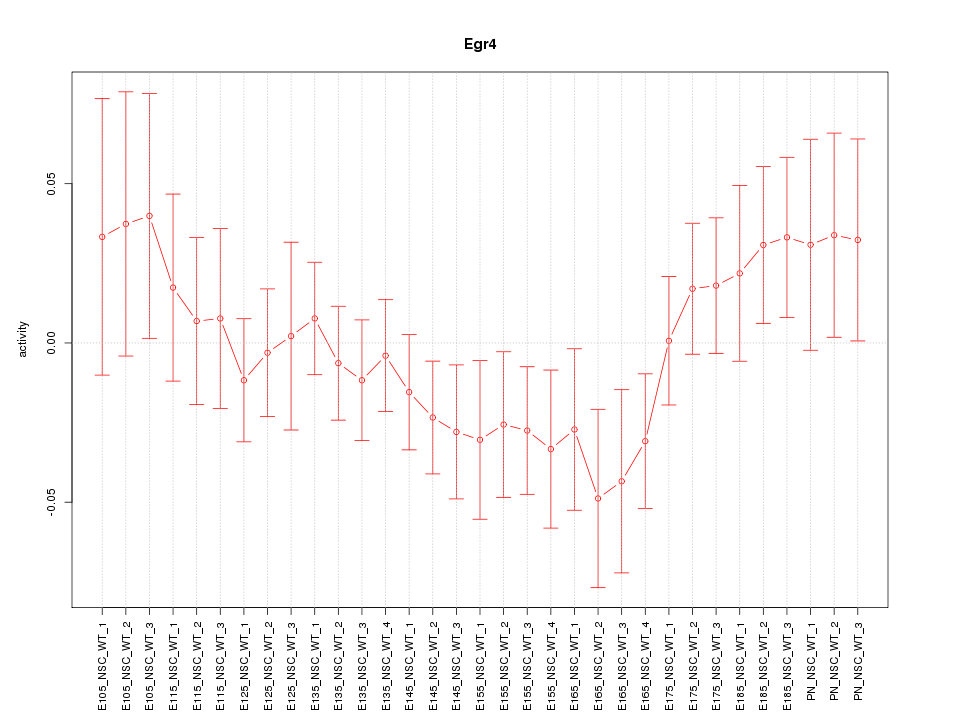
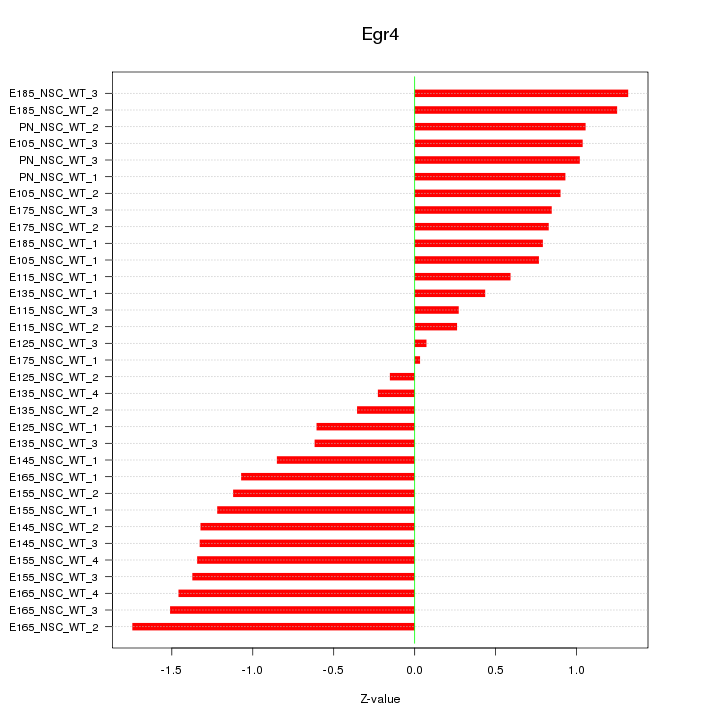
| 0.7 |
2.0 |
GO:0003100 |
angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) regulation of systemic arterial blood pressure by endothelin(GO:0003100) beta selection(GO:0043366) |
| 0.6 |
1.8 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.5 |
3.6 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.5 |
3.6 |
GO:0061304 |
retinal blood vessel morphogenesis(GO:0061304) |
| 0.5 |
1.5 |
GO:0010752 |
negative regulation of antigen processing and presentation(GO:0002578) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) negative regulation of plasminogen activation(GO:0010757) |
| 0.4 |
6.5 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 |
1.2 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.4 |
1.5 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.3 |
0.9 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 |
2.4 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.3 |
1.2 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 |
1.4 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 |
3.9 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.3 |
1.3 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) |
| 0.2 |
3.2 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 |
1.4 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.2 |
1.1 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 |
1.7 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.2 |
1.4 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 |
0.7 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.2 |
4.0 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.2 |
1.0 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.5 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) |
| 0.1 |
0.4 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
0.7 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.1 |
1.2 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 |
2.6 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 |
1.8 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.1 |
0.6 |
GO:1900016 |
negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.1 |
0.4 |
GO:0048319 |
axial mesoderm morphogenesis(GO:0048319) |
| 0.1 |
0.9 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.1 |
1.1 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.4 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.1 |
2.5 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.1 |
2.0 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
1.5 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.1 |
3.5 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 |
2.7 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
1.5 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
3.0 |
GO:0070613 |
regulation of protein processing(GO:0070613) |
| 0.0 |
3.9 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.5 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.5 |
GO:0010719 |
negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 |
0.1 |
GO:0044340 |
canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 |
0.1 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 |
0.6 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
1.4 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.3 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
1.1 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
1.3 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.8 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.6 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 |
0.2 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.0 |
4.0 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 |
3.0 |
GO:0006006 |
glucose metabolic process(GO:0006006) |
| 0.0 |
0.6 |
GO:2000463 |
regulation of long-term neuronal synaptic plasticity(GO:0048169) receptor localization to synapse(GO:0097120) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 |
0.5 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.5 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.0 |
1.6 |
GO:0043123 |
positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |