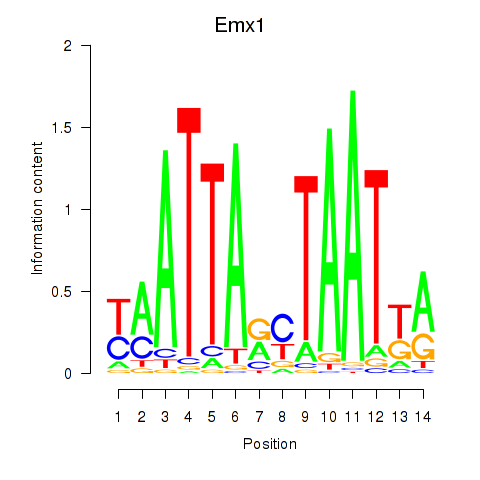
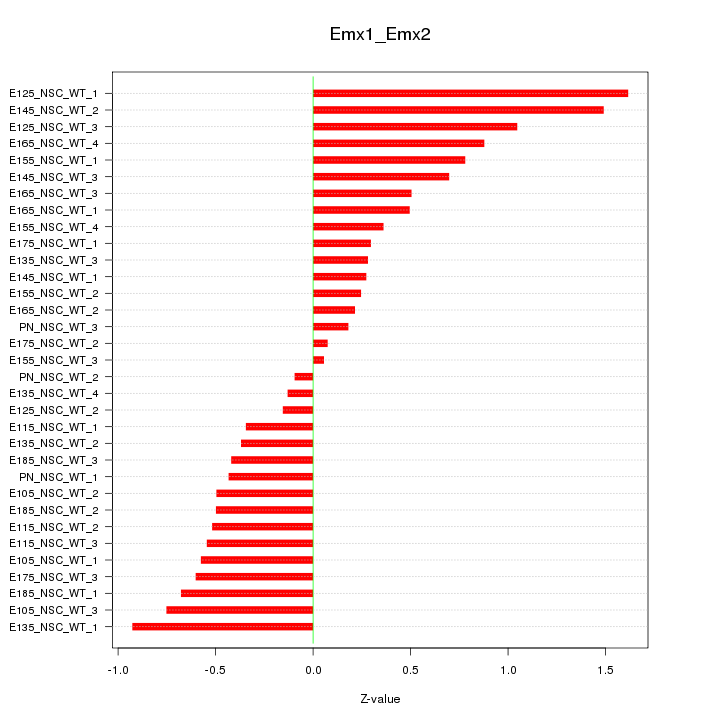
| 3.7 |
14.9 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 |
1.5 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.5 |
1.4 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.4 |
1.3 |
GO:2000564 |
CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.4 |
0.8 |
GO:0021730 |
trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.3 |
1.1 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.3 |
1.0 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.3 |
0.8 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 |
0.6 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.2 |
0.4 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.2 |
1.8 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.2 |
1.8 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 |
0.7 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.1 |
0.8 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.5 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
1.3 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
0.5 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 |
0.3 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 |
0.5 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.1 |
0.4 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.5 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 |
0.8 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.1 |
0.3 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 |
0.2 |
GO:0050787 |
glycolate metabolic process(GO:0009441) enzyme active site formation(GO:0018307) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) glyoxal catabolic process(GO:1903190) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 |
0.2 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 |
0.6 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 |
0.8 |
GO:0055059 |
asymmetric neuroblast division(GO:0055059) |
| 0.1 |
0.5 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.1 |
GO:0010519 |
negative regulation of phospholipase activity(GO:0010519) |
| 0.0 |
0.7 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
0.3 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.1 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 |
2.2 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
1.0 |
GO:0030262 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 |
0.4 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.1 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.2 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.1 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 |
1.6 |
GO:0038083 |
peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 |
0.1 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 |
0.3 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.4 |
GO:0061162 |
establishment of monopolar cell polarity(GO:0061162) |
| 0.0 |
0.1 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.3 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.2 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.1 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.7 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.2 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.4 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.0 |
GO:0051542 |
elastin biosynthetic process(GO:0051542) |
| 0.0 |
0.1 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
2.2 |
GO:0048469 |
cell maturation(GO:0048469) |
| 0.0 |
0.5 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.5 |
GO:0051926 |
negative regulation of calcium ion transport(GO:0051926) |
| 0.0 |
0.1 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |