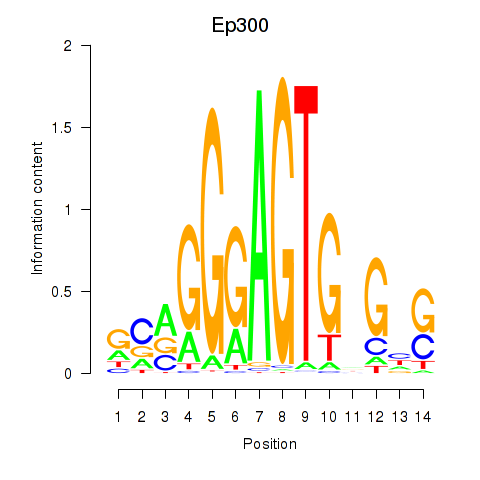
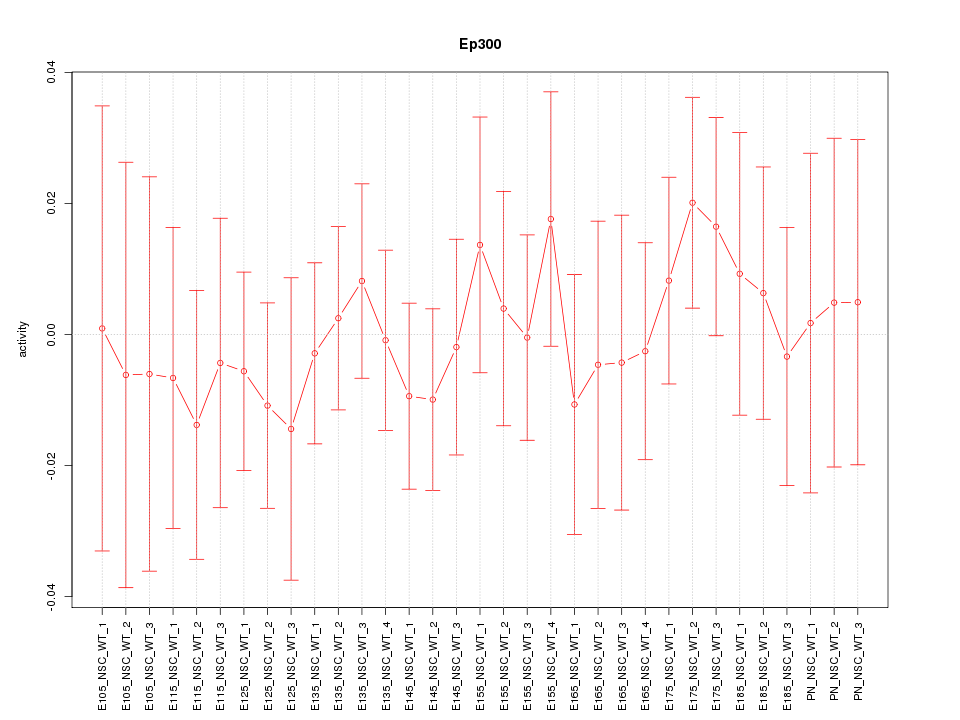
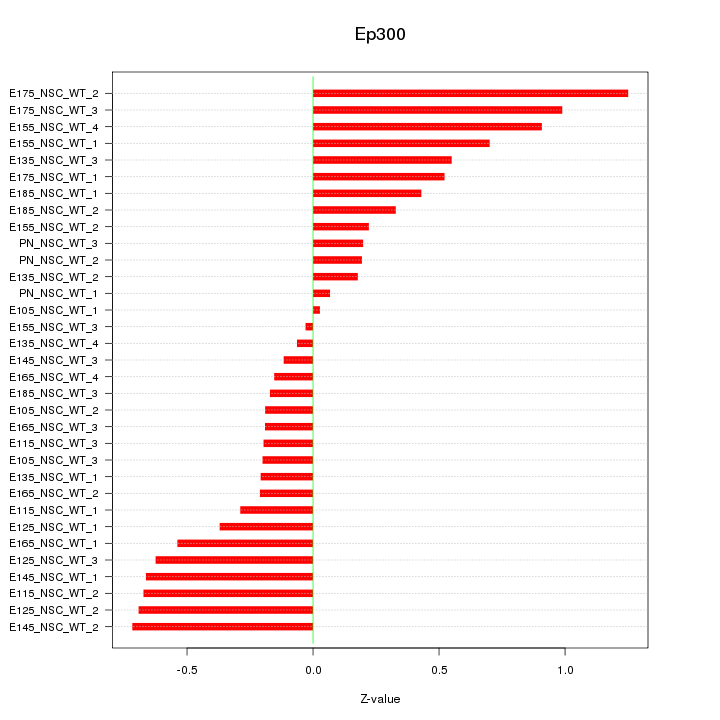
| 0.4 |
2.6 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
0.6 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.2 |
0.8 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.2 |
0.5 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) musculoskeletal movement, spinal reflex action(GO:0050883) olfactory pit development(GO:0060166) |
| 0.2 |
0.8 |
GO:0060279 |
positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.3 |
GO:0050713 |
negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 |
0.5 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 |
1.4 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.5 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
0.3 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 |
0.2 |
GO:2000564 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 |
0.2 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 |
0.2 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.1 |
0.8 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.0 |
0.1 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.0 |
0.5 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.6 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 |
0.2 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 |
0.3 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.3 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.7 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 0.0 |
0.3 |
GO:0032494 |
response to peptidoglycan(GO:0032494) |
| 0.0 |
0.3 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 |
0.3 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.5 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.3 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.3 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.2 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 |
0.4 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.4 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 |
0.1 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.1 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.1 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.0 |
0.1 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.1 |
GO:1904154 |
trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 |
0.3 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.0 |
0.1 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 |
0.2 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.0 |
0.1 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 |
1.4 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.8 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
0.1 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 |
0.1 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 |
0.3 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.9 |
GO:0001676 |
long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 |
1.6 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.0 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
0.4 |
GO:0010738 |
regulation of protein kinase A signaling(GO:0010738) |
| 0.0 |
0.7 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.1 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.0 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 |
0.1 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 |
0.0 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.4 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.1 |
GO:1903963 |
arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 |
0.0 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 |
0.1 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.1 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.1 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |