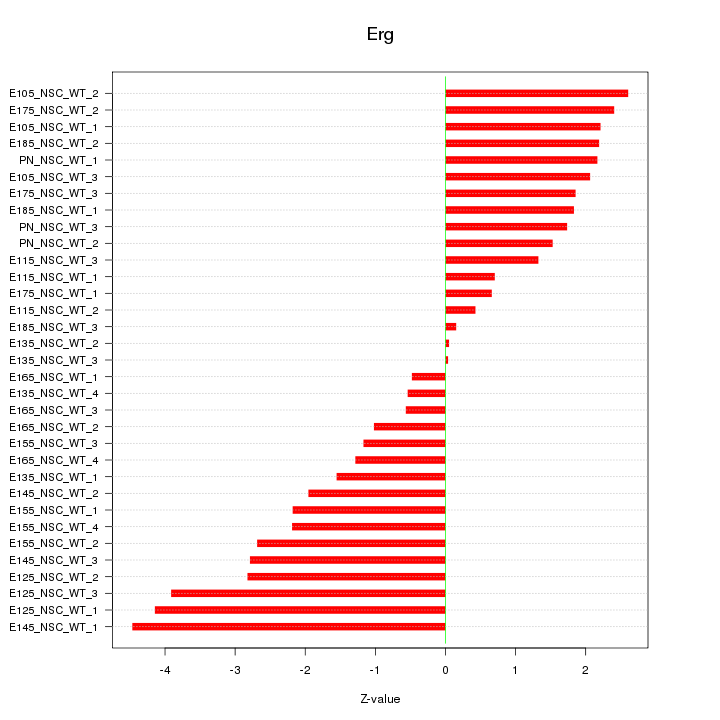
Motif ID: Erg
Z-value: 2.077

Transcription factors associated with Erg:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Erg | ENSMUSG00000040732.12 | Erg |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Erg | mm10_v2_chr16_-_95459245_95459384 | 0.48 | 5.0e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 3.9 | 11.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 2.9 | 8.6 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 2.4 | 7.3 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 2.4 | 12.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 2.2 | 6.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 2.0 | 7.9 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.8 | 5.3 | GO:0060437 | lung growth(GO:0060437) |
| 1.6 | 8.2 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 1.6 | 6.3 | GO:1904192 | prostate gland stromal morphogenesis(GO:0060741) cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 1.6 | 4.7 | GO:0003032 | detection of oxygen(GO:0003032) |
| 1.5 | 4.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.4 | 10.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 1.4 | 7.1 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 1.4 | 4.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.4 | 9.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.3 | 5.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 1.3 | 3.9 | GO:0090187 | zymogen granule exocytosis(GO:0070625) positive regulation of pancreatic juice secretion(GO:0090187) |
| 1.1 | 4.6 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 1.1 | 6.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.0 | 5.0 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 1.0 | 2.9 | GO:0042939 | renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.0 | 5.8 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 1.0 | 3.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.9 | 5.7 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.9 | 16.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.9 | 5.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.9 | 3.6 | GO:0050904 | diapedesis(GO:0050904) |
| 0.9 | 3.5 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.8 | 0.8 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.8 | 1.6 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.8 | 3.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.8 | 2.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.8 | 3.8 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.8 | 6.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.7 | 2.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.7 | 3.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.7 | 3.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.7 | 6.4 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.7 | 7.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.7 | 2.1 | GO:0035880 | cell proliferation in midbrain(GO:0033278) embryonic nail plate morphogenesis(GO:0035880) |
| 0.7 | 3.5 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.7 | 2.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.7 | 2.0 | GO:0046544 | regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) response to interleukin-15(GO:0070672) |
| 0.7 | 2.7 | GO:0042637 | catagen(GO:0042637) |
| 0.6 | 10.3 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.6 | 1.8 | GO:0060915 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.6 | 3.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.6 | 2.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.6 | 4.7 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.6 | 4.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.6 | 1.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.6 | 1.7 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.5 | 1.6 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.5 | 0.5 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.5 | 1.4 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.5 | 3.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.5 | 4.7 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.5 | 1.4 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.5 | 2.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.5 | 1.4 | GO:2000256 | positive regulation of male germ cell proliferation(GO:2000256) |
| 0.4 | 3.6 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.4 | 1.3 | GO:0021943 | calcium-independent cell-matrix adhesion(GO:0007161) formation of radial glial scaffolds(GO:0021943) |
| 0.4 | 1.8 | GO:0010878 | cholesterol storage(GO:0010878) |
| 0.4 | 1.8 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.4 | 2.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.4 | 4.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 2.5 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.4 | 1.6 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.4 | 4.8 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.4 | 3.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 3.2 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.4 | 7.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.4 | 12.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.4 | 1.9 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 2.6 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.4 | 0.7 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 | 7.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 0.7 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.3 | 3.8 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.3 | 3.8 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.3 | 2.7 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.3 | 1.0 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.3 | 2.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 2.0 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.3 | 2.4 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.3 | 1.4 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.3 | 1.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.3 | 2.0 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.3 | 2.0 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.3 | 12.1 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.3 | 3.8 | GO:0030238 | male sex determination(GO:0030238) |
| 0.3 | 1.6 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 | 2.8 | GO:2001053 | regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.3 | 1.5 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.3 | 18.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.3 | 6.0 | GO:0001946 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.3 | 2.0 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.3 | 0.9 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.3 | 0.8 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.3 | 1.9 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.3 | 1.1 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.3 | 1.9 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.3 | 2.5 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.3 | 1.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 2.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 0.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.2 | 3.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 2.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 2.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.2 | 0.9 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 0.7 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 | 0.4 | GO:0051462 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 0.2 | 1.6 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 2.4 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 1.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 0.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 0.4 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.2 | 2.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 0.5 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.2 | 2.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 1.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 1.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 0.5 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.2 | 3.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 4.1 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 0.9 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.4 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.4 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 1.8 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.1 | 0.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.7 | GO:0097049 | motor neuron apoptotic process(GO:0097049) regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 2.6 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.7 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 1.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.5 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 3.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 | 4.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 1.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 2.7 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.8 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 0.3 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.8 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 1.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.6 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 4.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.1 | 5.2 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 0.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.2 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) |
| 0.1 | 4.8 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 2.8 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.1 | 3.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 3.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.2 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 0.5 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 2.7 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.3 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 1.9 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 1.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 1.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 1.6 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 2.0 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 2.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.9 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 1.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-dependent nucleosome organization(GO:0034723) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.4 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 6.0 | GO:0014013 | regulation of gliogenesis(GO:0014013) |
| 0.1 | 0.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 1.6 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 0.6 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.7 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.8 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 4.5 | GO:0090132 | epithelial cell migration(GO:0010631) epithelium migration(GO:0090132) |
| 0.0 | 2.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.8 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.3 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 2.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.0 | 2.6 | GO:0006664 | glycolipid metabolic process(GO:0006664) liposaccharide metabolic process(GO:1903509) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.9 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 3.2 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 2.9 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 3.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 1.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 1.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.5 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 1.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.9 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 1.5 | 13.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.3 | 5.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.1 | 3.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.1 | 5.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.0 | 4.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 2.5 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.8 | 2.5 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.8 | 11.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.7 | 3.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.7 | 7.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.6 | 2.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 9.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 1.3 | GO:0034679 | integrin alpha2-beta1 complex(GO:0034666) integrin alpha3-beta1 complex(GO:0034667) integrin alpha9-beta1 complex(GO:0034679) |
| 0.4 | 4.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.4 | 3.0 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.4 | 1.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.4 | 10.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.4 | 1.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.4 | 2.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 17.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.3 | 3.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.3 | 2.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 6.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 22.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 4.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 5.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 2.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 4.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 5.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 2.0 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 6.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 15.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 1.3 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 0.9 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.2 | 3.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 0.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 7.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 14.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.4 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 1.7 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 20.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 2.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 4.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 3.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 12.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 6.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 14.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 5.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 4.7 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 2.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 4.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 3.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 2.8 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 4.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 3.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 3.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 8.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 6.0 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 2.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 17.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 4.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 7.1 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 2.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:0098858 | actin-based cell projection(GO:0098858) |
| 0.0 | 1.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 2.0 | 12.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 2.0 | 7.9 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.6 | 4.7 | GO:0005534 | galactose binding(GO:0005534) |
| 1.5 | 6.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 1.2 | 13.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.2 | 20.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 1.1 | 3.4 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 1.1 | 7.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.9 | 3.8 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.8 | 4.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.8 | 2.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.7 | 11.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.7 | 2.8 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.7 | 3.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.7 | 2.7 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.7 | 2.0 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.7 | 2.7 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.7 | 15.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.7 | 2.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.6 | 3.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.6 | 2.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.6 | 9.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.6 | 4.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.6 | 2.4 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.6 | 1.7 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.6 | 6.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.5 | 1.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.5 | 1.5 | GO:0008521 | acetyl-CoA transporter activity(GO:0008521) |
| 0.5 | 1.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 3.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.5 | 1.9 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.5 | 8.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.5 | 4.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.4 | 3.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.4 | 1.7 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.4 | 1.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 16.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.4 | 2.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 1.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 8.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 3.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 4.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 2.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 2.0 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.3 | 1.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.3 | 1.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 11.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 1.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 4.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 2.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 1.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.7 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 3.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 2.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 7.4 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.2 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 2.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.2 | 6.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 3.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.5 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.2 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 11.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 20.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 6.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 4.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.7 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 3.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 9.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 7.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.7 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.1 | 2.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 2.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.5 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 3.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.7 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 0.9 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.8 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 17.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 13.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 8.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 4.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 7.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.8 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) |
| 0.1 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 4.7 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 10.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 2.3 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 2.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 7.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.1 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 14.1 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 3.3 | GO:0000975 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.9 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 2.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.9 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.4 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.9 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |