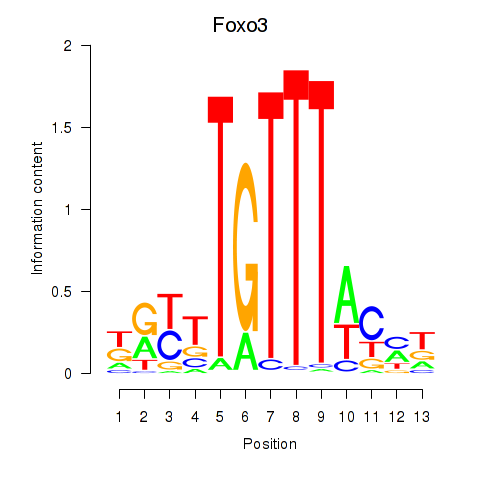
Motif ID: Foxo3
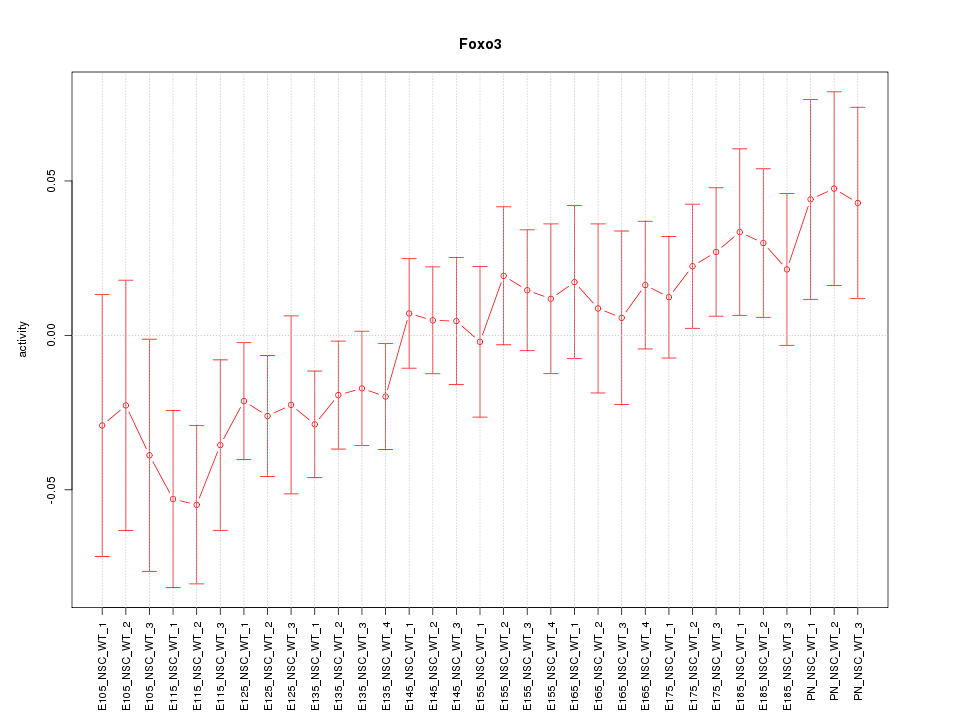
Z-value: 1.068
Transcription factors associated with Foxo3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxo3 | ENSMUSG00000048756.5 | Foxo3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo3 | mm10_v2_chr10_-_42276744_42276763 | 0.81 | 9.9e-09 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:1902524 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 1.1 | 15.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.1 | 5.4 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 1.0 | 7.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.9 | 4.7 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.8 | 2.3 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.7 | 4.8 | GO:1903726 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.5 | 8.4 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.5 | 14.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.4 | 3.2 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.3 | 2.0 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.3 | 10.4 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.2 | 2.0 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.2 | 0.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 1.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.7 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.7 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 | 1.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.0 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 2.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.5 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.9 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 1.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 2.0 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.3 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.1 | 0.5 | GO:0015791 | polyol transport(GO:0015791) urea transport(GO:0015840) bile acid secretion(GO:0032782) urea transmembrane transport(GO:0071918) |
| 0.1 | 3.9 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.1 | 1.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 2.8 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.4 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 1.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.2 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 4.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.8 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.2 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.2 | 1.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 2.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 3.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 3.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 5.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 3.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 30.3 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 8.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.6 | 4.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.3 | 5.4 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 1.3 | 5.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.7 | 2.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.7 | 2.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.6 | 2.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 4.7 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.4 | 14.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.7 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 1.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 3.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 3.9 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.5 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 2.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 15.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 1.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 2.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.0 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |