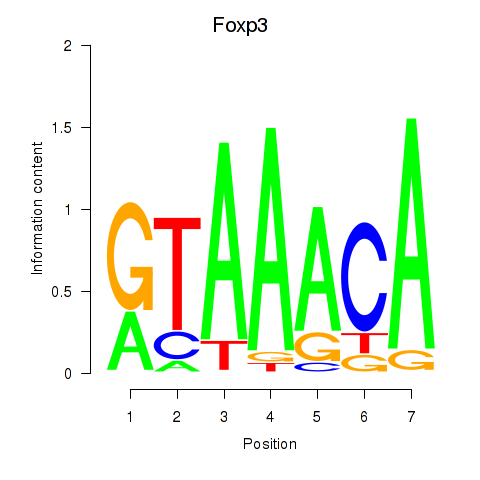
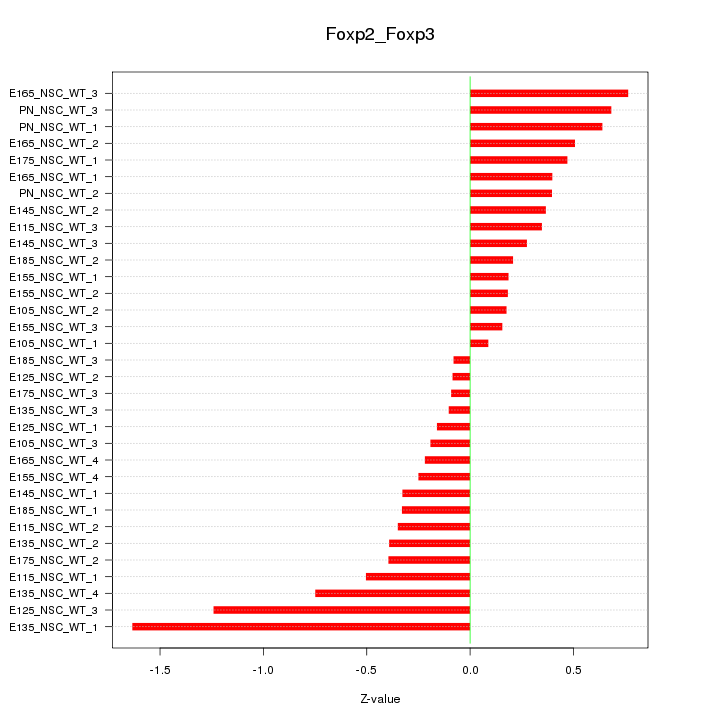
| 0.5 |
2.6 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.5 |
5.5 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.5 |
1.4 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.4 |
2.7 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.4 |
1.1 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.3 |
1.0 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 |
1.2 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 |
1.5 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 |
1.2 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.3 |
0.9 |
GO:0090403 |
oxidative stress-induced premature senescence(GO:0090403) |
| 0.3 |
1.2 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 |
0.8 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.3 |
0.8 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.3 |
1.8 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.2 |
0.7 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 |
1.6 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.2 |
1.6 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.2 |
0.7 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 |
0.5 |
GO:1900623 |
regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 |
1.8 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.2 |
2.2 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.2 |
0.9 |
GO:0055099 |
regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 0.2 |
1.1 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.2 |
1.0 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 |
0.2 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.2 |
0.6 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.2 |
0.8 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.2 |
2.5 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.2 |
0.7 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 |
0.9 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 |
0.5 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.2 |
0.3 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 |
1.2 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 |
1.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 |
1.6 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.2 |
2.7 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.2 |
0.5 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.2 |
0.5 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.2 |
0.3 |
GO:0010751 |
negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 |
1.4 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
1.2 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.1 |
0.4 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 |
0.4 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.9 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
1.0 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
0.8 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.2 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 |
0.4 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.1 |
1.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 |
0.5 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
2.0 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 |
0.5 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 |
1.4 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.4 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.1 |
0.8 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.1 |
1.4 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 |
0.3 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.1 |
0.3 |
GO:1902219 |
maintenance of blood-brain barrier(GO:0035633) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
0.2 |
GO:0070487 |
monocyte aggregation(GO:0070487) |
| 0.1 |
0.9 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
0.2 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.4 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 |
0.4 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.1 |
0.7 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 |
0.2 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 |
0.4 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.2 |
GO:2000598 |
regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 |
0.3 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.1 |
0.1 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.1 |
0.3 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.1 |
0.3 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.4 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 |
0.3 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 |
0.3 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.1 |
1.0 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 |
0.7 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.6 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 |
0.4 |
GO:0002904 |
positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 |
0.5 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.3 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 |
0.2 |
GO:1901894 |
regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.1 |
0.1 |
GO:2000481 |
positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 |
1.0 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.2 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 |
0.4 |
GO:0097460 |
ferrous iron import into cell(GO:0097460) |
| 0.0 |
0.2 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 |
0.4 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.0 |
0.3 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.0 |
0.3 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 |
0.2 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 |
0.1 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of cyclase activity(GO:0031280) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 |
0.1 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.0 |
0.5 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.2 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
0.9 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.4 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.2 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.1 |
GO:1903054 |
negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 |
0.1 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.0 |
0.3 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 |
0.1 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 |
0.2 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.4 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 |
0.1 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.0 |
0.2 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.5 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.7 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
0.1 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.6 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 |
0.2 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.3 |
GO:0036120 |
response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 |
0.2 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.2 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.1 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.0 |
0.1 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 |
0.2 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 |
0.6 |
GO:0043547 |
positive regulation of GTPase activity(GO:0043547) |
| 0.0 |
0.1 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.0 |
0.2 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.0 |
0.5 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 |
0.4 |
GO:0061157 |
mRNA destabilization(GO:0061157) |
| 0.0 |
0.2 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.0 |
0.1 |
GO:0042368 |
negative regulation of vitamin D biosynthetic process(GO:0010957) vitamin D biosynthetic process(GO:0042368) negative regulation of vitamin metabolic process(GO:0046137) regulation of vitamin D biosynthetic process(GO:0060556) regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
0.5 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.1 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 |
1.5 |
GO:0010828 |
positive regulation of glucose transport(GO:0010828) |
| 0.0 |
0.1 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.0 |
0.2 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.0 |
0.3 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.0 |
0.3 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.0 |
0.4 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.3 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.0 |
0.9 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.2 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 |
0.1 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.0 |
0.1 |
GO:0071635 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 |
0.5 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.0 |
0.2 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.5 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 |
0.2 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 |
0.4 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.4 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.3 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.1 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 |
0.8 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.1 |
GO:0050929 |
corticospinal neuron axon guidance(GO:0021966) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 |
0.4 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.7 |
GO:0048662 |
negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 |
0.1 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.4 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.1 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.3 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.1 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 |
0.7 |
GO:0060612 |
adipose tissue development(GO:0060612) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.1 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.3 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.1 |
GO:0002755 |
MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 |
0.4 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.5 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.2 |
GO:0051302 |
regulation of cell division(GO:0051302) |
| 0.0 |
0.5 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.2 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.2 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.1 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.7 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.5 |
GO:1900449 |
regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 |
0.4 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.2 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
0.1 |
GO:1903056 |
melanocyte migration(GO:0097324) regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 |
0.2 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.1 |
GO:0015884 |
folic acid transport(GO:0015884) |
| 0.0 |
0.4 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 |
0.3 |
GO:0014741 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 |
0.3 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 |
0.2 |
GO:0003417 |
growth plate cartilage development(GO:0003417) |
| 0.0 |
0.2 |
GO:0072673 |
lamellipodium morphogenesis(GO:0072673) |
| 0.0 |
0.1 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 |
0.3 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.2 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.1 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 |
0.6 |
GO:0000271 |
polysaccharide biosynthetic process(GO:0000271) |
| 0.0 |
0.1 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.1 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
0.0 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.0 |
0.2 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.3 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.1 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |