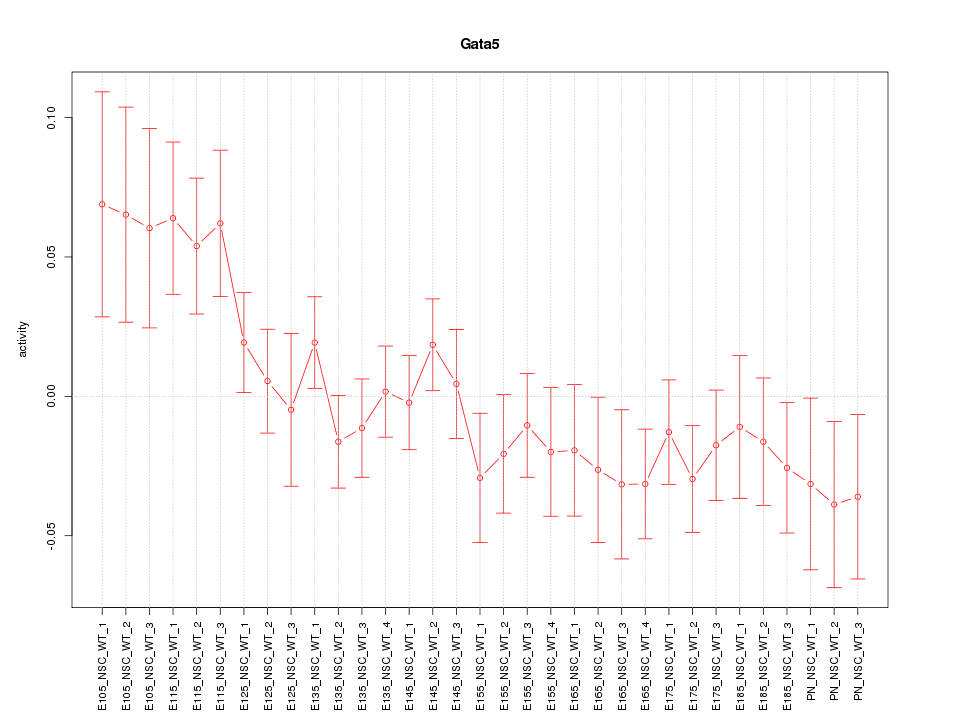
Motif ID: Gata5
Z-value: 1.218
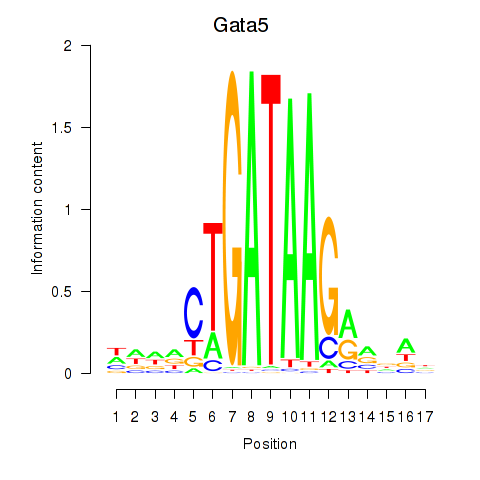
Transcription factors associated with Gata5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Gata5 | ENSMUSG00000015627.5 | Gata5 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.8 | 83.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.4 | 2.4 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 2.1 | 18.6 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.9 | 4.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.7 | 4.8 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.7 | 2.6 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.7 | 3.9 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.6 | 1.8 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.5 | 2.5 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.4 | 1.3 | GO:0046098 | purine nucleobase salvage(GO:0043096) guanine metabolic process(GO:0046098) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.4 | 3.9 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.4 | 1.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.4 | 6.3 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.3 | 1.1 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 0.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.7 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 1.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.9 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 1.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 1.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 4.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 | 0.7 | GO:0016068 | regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) type I hypersensitivity(GO:0016068) |
| 0.2 | 7.3 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 1.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.5 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.8 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 3.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.2 | GO:0032499 | detection of peptidoglycan(GO:0032499) |
| 0.1 | 0.8 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.3 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 1.9 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 0.5 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.1 | 0.5 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.4 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 1.1 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.5 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.1 | 3.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 3.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.0 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 2.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) membrane fission(GO:0090148) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 5.2 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.9 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 4.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.5 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 3.5 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 2.7 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 1.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.3 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.7 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.8 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.1 | GO:1903225 | regulation of endodermal cell fate specification(GO:0042663) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 7.8 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 0.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.4 | GO:0048806 | genitalia development(GO:0048806) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 3.9 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.4 | GO:0042755 | eating behavior(GO:0042755) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 91.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.6 | 4.8 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.6 | 2.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 1.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 2.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.3 | 0.8 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 4.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 2.1 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 4.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 1.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.7 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 7.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 3.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.5 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.8 | 83.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 5.8 | 17.3 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.9 | 7.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 1.3 | 3.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 2.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.6 | 1.8 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.6 | 3.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.5 | 1.6 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.5 | 2.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 1.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.4 | 6.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 1.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 4.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 1.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 2.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 1.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 4.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.3 | 0.8 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.3 | 1.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 1.9 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.2 | 1.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 3.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 3.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0070891 | peptidoglycan binding(GO:0042834) lipoteichoic acid binding(GO:0070891) |
| 0.0 | 3.2 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 2.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 3.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.5 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 2.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 2.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 3.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 1.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |