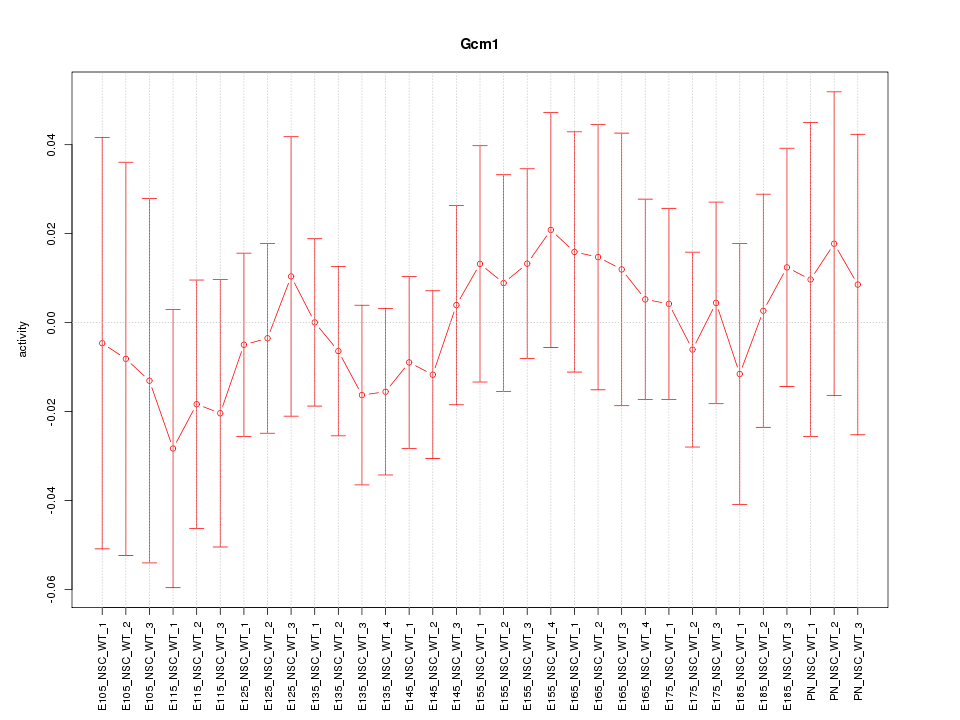
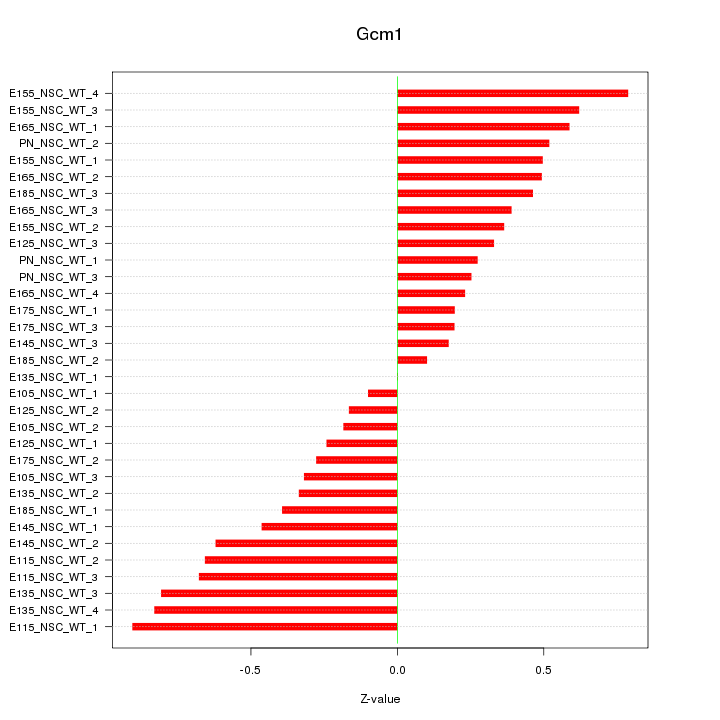
| 0.3 |
0.8 |
GO:0031686 |
A1 adenosine receptor binding(GO:0031686) |
| 0.2 |
0.8 |
GO:0038049 |
glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 |
0.6 |
GO:0030144 |
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 |
0.3 |
GO:0048019 |
receptor antagonist activity(GO:0048019) |
| 0.1 |
0.6 |
GO:0035381 |
extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) mercury ion binding(GO:0045340) |
| 0.1 |
2.4 |
GO:0043274 |
phospholipase binding(GO:0043274) |
| 0.1 |
0.6 |
GO:0005004 |
GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 |
1.9 |
GO:0005251 |
delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 |
0.3 |
GO:0016176 |
superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 |
0.8 |
GO:0005388 |
calcium-transporting ATPase activity(GO:0005388) |
| 0.1 |
0.9 |
GO:0004000 |
adenosine deaminase activity(GO:0004000) |
| 0.1 |
1.0 |
GO:0031681 |
G-protein beta-subunit binding(GO:0031681) |
| 0.1 |
0.4 |
GO:0008499 |
UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 |
1.0 |
GO:0005545 |
1-phosphatidylinositol binding(GO:0005545) |
| 0.0 |
0.1 |
GO:0003846 |
2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 |
0.2 |
GO:0017002 |
activin-activated receptor activity(GO:0017002) |
| 0.0 |
0.4 |
GO:0015026 |
coreceptor activity(GO:0015026) |
| 0.0 |
0.4 |
GO:1990841 |
promoter-specific chromatin binding(GO:1990841) |
| 0.0 |
0.2 |
GO:0015643 |
toxic substance binding(GO:0015643) |
| 0.0 |
0.3 |
GO:0015095 |
magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 |
0.5 |
GO:0034945 |
dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 |
0.1 |
GO:0061133 |
endopeptidase activator activity(GO:0061133) |