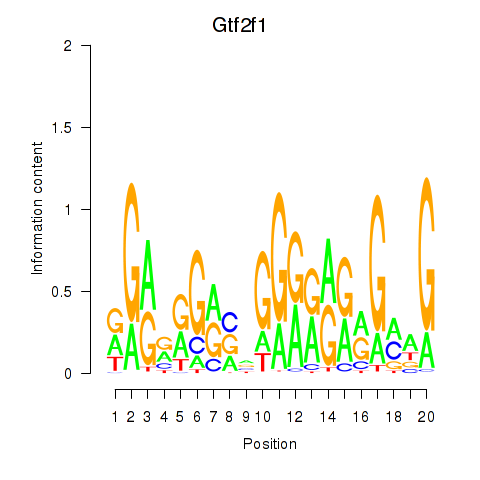
| 13.1 |
39.2 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 7.6 |
30.5 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 7.5 |
22.5 |
GO:0046959 |
habituation(GO:0046959) |
| 6.1 |
18.3 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 5.7 |
34.0 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 4.9 |
19.8 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 4.5 |
13.6 |
GO:0060023 |
soft palate development(GO:0060023) |
| 4.4 |
13.1 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 3.9 |
19.6 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 3.8 |
3.8 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 3.8 |
7.6 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 3.8 |
37.9 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 3.6 |
25.2 |
GO:0032811 |
negative regulation of epinephrine secretion(GO:0032811) |
| 3.6 |
10.8 |
GO:0046881 |
positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 3.4 |
53.6 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 3.2 |
9.5 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 3.2 |
38.1 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 2.9 |
17.6 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 2.9 |
8.7 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) |
| 2.9 |
8.7 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.8 |
14.1 |
GO:0031133 |
regulation of axon diameter(GO:0031133) |
| 2.8 |
8.4 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 2.8 |
8.3 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.7 |
10.9 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 2.6 |
15.9 |
GO:0032229 |
negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 2.6 |
41.5 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 2.6 |
10.2 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 2.5 |
12.7 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 2.5 |
25.1 |
GO:0010807 |
regulation of synaptic vesicle priming(GO:0010807) |
| 2.3 |
9.2 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 2.3 |
6.9 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 2.2 |
6.7 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 2.2 |
6.6 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 2.2 |
4.3 |
GO:0071205 |
protein localization to juxtaparanode region of axon(GO:0071205) |
| 2.1 |
8.4 |
GO:1901204 |
regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 2.1 |
6.2 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 2.0 |
9.9 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 1.9 |
7.8 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 1.9 |
3.8 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 1.9 |
22.9 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 1.8 |
5.3 |
GO:1904685 |
positive regulation of metalloendopeptidase activity(GO:1904685) |
| 1.7 |
5.2 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 1.7 |
5.1 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 1.7 |
15.2 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 1.7 |
8.4 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.6 |
3.2 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 1.6 |
19.2 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 1.6 |
8.0 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 1.6 |
7.8 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 1.6 |
1.6 |
GO:0034183 |
negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 1.6 |
12.5 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 1.6 |
4.7 |
GO:0060466 |
monocyte extravasation(GO:0035696) activation of meiosis involved in egg activation(GO:0060466) positive regulation of acrosome reaction(GO:2000344) regulation of monocyte extravasation(GO:2000437) |
| 1.5 |
4.6 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 1.5 |
7.7 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 1.5 |
10.7 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 1.5 |
4.5 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.5 |
6.0 |
GO:0038026 |
reelin-mediated signaling pathway(GO:0038026) |
| 1.5 |
7.4 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 1.5 |
7.3 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 1.4 |
8.7 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 1.4 |
7.1 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 1.4 |
2.8 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 1.4 |
22.1 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 1.3 |
6.7 |
GO:1904861 |
excitatory synapse assembly(GO:1904861) |
| 1.3 |
5.3 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.3 |
4.0 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 1.3 |
14.5 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 1.3 |
1.3 |
GO:0061669 |
spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.3 |
7.9 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 1.3 |
1.3 |
GO:0055005 |
ventricular cardiac myofibril assembly(GO:0055005) |
| 1.3 |
11.7 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 1.2 |
2.5 |
GO:1903421 |
regulation of synaptic vesicle recycling(GO:1903421) |
| 1.2 |
8.6 |
GO:0061502 |
early endosome to recycling endosome transport(GO:0061502) |
| 1.2 |
3.7 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.2 |
4.8 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 1.2 |
12.1 |
GO:2000463 |
positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 1.2 |
10.7 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.2 |
3.5 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 1.2 |
3.5 |
GO:0061341 |
non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 1.1 |
3.4 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 1.1 |
11.3 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 1.1 |
6.7 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 1.1 |
9.0 |
GO:0090394 |
negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 1.1 |
4.5 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 1.1 |
5.6 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.1 |
6.6 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 1.1 |
18.6 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 1.1 |
1.1 |
GO:0061535 |
glutamate secretion, neurotransmission(GO:0061535) |
| 1.1 |
4.3 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 1.1 |
4.3 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 1.1 |
5.4 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 1.1 |
13.8 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 1.0 |
3.1 |
GO:0021933 |
cerebellar cortex maturation(GO:0021699) radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 1.0 |
4.1 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.0 |
4.1 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.0 |
21.3 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 1.0 |
4.0 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) |
| 1.0 |
5.0 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 1.0 |
7.7 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 1.0 |
3.9 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 1.0 |
4.8 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 1.0 |
2.9 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.0 |
5.7 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 1.0 |
1.0 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.9 |
0.9 |
GO:0046958 |
nonassociative learning(GO:0046958) |
| 0.9 |
3.7 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.9 |
2.8 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.9 |
7.3 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.9 |
4.5 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.9 |
4.5 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.9 |
0.9 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.9 |
0.9 |
GO:0055119 |
relaxation of cardiac muscle(GO:0055119) |
| 0.9 |
4.4 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.9 |
24.5 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.9 |
16.5 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.9 |
0.9 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.8 |
9.2 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.8 |
5.0 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.8 |
3.9 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.8 |
3.1 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) peptidyl-aspartic acid modification(GO:0018197) |
| 0.8 |
16.1 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.8 |
4.6 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.8 |
2.3 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.8 |
2.3 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.7 |
3.7 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.7 |
4.4 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.7 |
1.5 |
GO:0097051 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.7 |
2.9 |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.7 |
2.2 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.7 |
2.9 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.7 |
6.6 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.7 |
2.2 |
GO:0007403 |
glial cell fate determination(GO:0007403) |
| 0.7 |
8.6 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.7 |
4.3 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.7 |
2.1 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.7 |
16.3 |
GO:0022401 |
desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.7 |
1.4 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.7 |
2.1 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.7 |
4.2 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.7 |
1.4 |
GO:0035898 |
parathyroid hormone secretion(GO:0035898) |
| 0.7 |
0.7 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.7 |
3.5 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.7 |
2.1 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.7 |
2.1 |
GO:0001927 |
exocyst assembly(GO:0001927) |
| 0.7 |
3.4 |
GO:2000786 |
positive regulation of vacuole organization(GO:0044090) positive regulation of autophagosome assembly(GO:2000786) |
| 0.7 |
4.8 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.7 |
9.4 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.7 |
10.1 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.7 |
0.7 |
GO:0098735 |
positive regulation of the force of heart contraction(GO:0098735) |
| 0.7 |
2.7 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.7 |
1.3 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.7 |
0.7 |
GO:0051835 |
positive regulation of synapse structural plasticity(GO:0051835) |
| 0.7 |
2.0 |
GO:0033024 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.7 |
2.0 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.7 |
4.6 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.7 |
1.3 |
GO:2001023 |
regulation of response to drug(GO:2001023) |
| 0.6 |
1.3 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.6 |
3.2 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.6 |
1.9 |
GO:0060453 |
regulation of gastric acid secretion(GO:0060453) |
| 0.6 |
3.2 |
GO:0048520 |
positive regulation of behavior(GO:0048520) |
| 0.6 |
2.5 |
GO:2000272 |
negative regulation of receptor activity(GO:2000272) |
| 0.6 |
2.5 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.6 |
2.5 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.6 |
1.9 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.6 |
1.3 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.6 |
16.9 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.6 |
1.9 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.6 |
15.5 |
GO:0035640 |
exploration behavior(GO:0035640) |
| 0.6 |
3.7 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.6 |
0.6 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.6 |
7.1 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.6 |
4.1 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.6 |
2.9 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.6 |
4.6 |
GO:0008038 |
neuron recognition(GO:0008038) |
| 0.6 |
4.6 |
GO:0021540 |
corpus callosum morphogenesis(GO:0021540) |
| 0.6 |
1.1 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.6 |
13.2 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.6 |
1.7 |
GO:0014809 |
regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.6 |
2.3 |
GO:1902669 |
positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.6 |
2.8 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.6 |
1.7 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.6 |
6.7 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.6 |
1.7 |
GO:0043313 |
regulation of neutrophil degranulation(GO:0043313) |
| 0.6 |
5.0 |
GO:0051195 |
negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.6 |
2.2 |
GO:0050812 |
regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.5 |
9.3 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.5 |
3.8 |
GO:0014824 |
artery smooth muscle contraction(GO:0014824) |
| 0.5 |
5.4 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.5 |
1.1 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.5 |
2.1 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.5 |
2.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.5 |
2.7 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.5 |
2.7 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.5 |
3.2 |
GO:0060509 |
Type I pneumocyte differentiation(GO:0060509) |
| 0.5 |
1.6 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.5 |
17.4 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.5 |
29.4 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.5 |
3.1 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.5 |
3.1 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.5 |
3.6 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) |
| 0.5 |
6.1 |
GO:0010832 |
negative regulation of myotube differentiation(GO:0010832) |
| 0.5 |
1.5 |
GO:0045404 |
positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.5 |
6.0 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.5 |
1.0 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.5 |
1.5 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.5 |
4.3 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 |
2.4 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.5 |
1.4 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.5 |
2.4 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.5 |
2.3 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.5 |
1.4 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.5 |
4.2 |
GO:0098907 |
T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.5 |
1.4 |
GO:0033364 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.5 |
1.4 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.5 |
1.8 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.5 |
9.6 |
GO:0060292 |
long term synaptic depression(GO:0060292) |
| 0.5 |
3.7 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.5 |
0.9 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.5 |
0.9 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.4 |
4.0 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.4 |
1.8 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.4 |
1.8 |
GO:0051966 |
regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.4 |
2.2 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.4 |
2.2 |
GO:0034141 |
positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.4 |
2.6 |
GO:0060907 |
positive regulation of macrophage cytokine production(GO:0060907) |
| 0.4 |
4.8 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 |
2.6 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.4 |
0.4 |
GO:0014722 |
regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.4 |
2.2 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.4 |
3.0 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.4 |
2.1 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.4 |
0.9 |
GO:0097252 |
oligodendrocyte apoptotic process(GO:0097252) |
| 0.4 |
2.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.4 |
17.8 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.4 |
1.7 |
GO:0071415 |
cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.4 |
2.1 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.4 |
2.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.4 |
1.7 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.4 |
21.7 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.4 |
2.5 |
GO:0089711 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) L-glutamate transmembrane transport(GO:0089711) |
| 0.4 |
1.2 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 |
2.9 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 |
2.5 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.4 |
0.8 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.4 |
3.6 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.4 |
1.2 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.4 |
3.6 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.4 |
1.6 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.4 |
0.4 |
GO:0045342 |
MHC class II biosynthetic process(GO:0045342) regulation of MHC class II biosynthetic process(GO:0045346) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 |
2.3 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.4 |
1.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.4 |
2.2 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.4 |
3.0 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.4 |
1.5 |
GO:0006538 |
glutamate catabolic process(GO:0006538) neurotransmitter biosynthetic process(GO:0042136) |
| 0.4 |
1.1 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 0.4 |
34.0 |
GO:0007612 |
learning(GO:0007612) |
| 0.4 |
0.4 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.4 |
1.1 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.4 |
3.6 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.4 |
24.2 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.4 |
1.1 |
GO:2000015 |
regulation of determination of dorsal identity(GO:2000015) |
| 0.4 |
1.1 |
GO:0048505 |
regulation of timing of cell differentiation(GO:0048505) |
| 0.4 |
2.5 |
GO:0002524 |
hypersensitivity(GO:0002524) |
| 0.4 |
2.5 |
GO:0055059 |
asymmetric neuroblast division(GO:0055059) |
| 0.4 |
1.8 |
GO:0097343 |
ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.3 |
0.3 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.3 |
2.4 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 |
1.0 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 |
1.7 |
GO:0032095 |
regulation of response to food(GO:0032095) negative regulation of response to food(GO:0032096) regulation of appetite(GO:0032098) negative regulation of appetite(GO:0032099) |
| 0.3 |
6.6 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 |
2.6 |
GO:0015844 |
monoamine transport(GO:0015844) |
| 0.3 |
0.7 |
GO:0036015 |
response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.3 |
1.0 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 |
2.9 |
GO:0032793 |
positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 |
0.3 |
GO:0034241 |
positive regulation of macrophage fusion(GO:0034241) |
| 0.3 |
0.9 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.3 |
1.3 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.3 |
1.6 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.3 |
5.9 |
GO:0099601 |
regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.3 |
0.6 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.3 |
1.8 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.3 |
20.8 |
GO:0006836 |
neurotransmitter transport(GO:0006836) |
| 0.3 |
2.7 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 |
1.5 |
GO:0019346 |
homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.3 |
0.9 |
GO:0097503 |
sialylation(GO:0097503) protein sialylation(GO:1990743) |
| 0.3 |
0.3 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.3 |
0.9 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.3 |
1.5 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.3 |
2.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.3 |
1.5 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.3 |
0.9 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 |
9.8 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.3 |
0.9 |
GO:0061386 |
closure of optic fissure(GO:0061386) |
| 0.3 |
2.0 |
GO:0006855 |
drug transmembrane transport(GO:0006855) |
| 0.3 |
0.3 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.3 |
0.3 |
GO:0002925 |
positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.3 |
6.2 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.3 |
1.7 |
GO:0071435 |
potassium ion export(GO:0071435) |
| 0.3 |
9.3 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 |
0.8 |
GO:0061418 |
regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.3 |
0.8 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.3 |
0.3 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.3 |
2.2 |
GO:0070445 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.3 |
1.9 |
GO:0015919 |
peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.3 |
10.2 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.3 |
2.4 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.3 |
1.3 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.3 |
0.8 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.3 |
3.2 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.3 |
2.6 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 |
7.4 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.3 |
1.3 |
GO:0009247 |
glycolipid biosynthetic process(GO:0009247) |
| 0.3 |
1.5 |
GO:0039535 |
regulation of RIG-I signaling pathway(GO:0039535) positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.3 |
4.1 |
GO:1902259 |
regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.3 |
1.3 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.3 |
0.8 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.3 |
3.8 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 |
1.3 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 |
0.8 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 |
2.5 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.2 |
1.0 |
GO:0001922 |
B-1 B cell homeostasis(GO:0001922) |
| 0.2 |
1.2 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 |
2.0 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 |
9.8 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.2 |
0.2 |
GO:1902837 |
amino acid import into cell(GO:1902837) |
| 0.2 |
1.5 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.2 |
0.2 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 |
0.5 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 |
5.5 |
GO:1901381 |
positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.2 |
8.4 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.2 |
1.0 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 |
1.7 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.2 |
0.7 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.2 |
0.7 |
GO:0060468 |
prevention of polyspermy(GO:0060468) |
| 0.2 |
3.2 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.2 |
1.1 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.2 |
0.2 |
GO:0010616 |
negative regulation of cardiac muscle adaptation(GO:0010616) |
| 0.2 |
1.8 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.2 |
0.9 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.2 |
0.5 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.2 |
4.3 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.2 |
4.1 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.2 |
0.7 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 |
3.1 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.2 |
0.9 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.2 |
0.4 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 |
0.6 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.2 |
0.9 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.2 |
1.1 |
GO:0030819 |
positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.2 |
0.4 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.2 |
0.6 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.2 |
1.9 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 |
1.7 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.2 |
1.1 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.2 |
1.7 |
GO:0090331 |
negative regulation of platelet aggregation(GO:0090331) |
| 0.2 |
0.8 |
GO:0044337 |
canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.2 |
0.6 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.2 |
0.2 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.2 |
3.4 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.2 |
3.8 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.2 |
0.6 |
GO:0071376 |
response to corticotropin-releasing hormone(GO:0043435) general adaptation syndrome(GO:0051866) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 |
1.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.2 |
1.6 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.2 |
0.6 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.2 |
0.8 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 |
0.8 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 |
1.1 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.2 |
2.1 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.2 |
0.4 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.2 |
0.8 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 |
0.4 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 |
0.6 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 |
0.4 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 |
0.9 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 |
5.9 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 |
0.5 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 |
3.4 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.2 |
9.7 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 |
1.6 |
GO:0048012 |
hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.2 |
1.4 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.2 |
1.4 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.2 |
1.4 |
GO:0051533 |
positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 |
2.6 |
GO:0036065 |
fucosylation(GO:0036065) |
| 0.2 |
2.4 |
GO:0045806 |
negative regulation of endocytosis(GO:0045806) |
| 0.2 |
0.2 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 |
1.7 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.2 |
2.4 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.2 |
1.0 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 |
3.5 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 |
0.8 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 |
1.7 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.2 |
0.7 |
GO:0097039 |
protein linear polyubiquitination(GO:0097039) |
| 0.2 |
11.5 |
GO:0030010 |
establishment of cell polarity(GO:0030010) |
| 0.2 |
4.3 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.2 |
1.6 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 |
0.3 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.2 |
1.0 |
GO:0061718 |
NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glycolytic process through glucose-1-phosphate(GO:0061622) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 |
0.6 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.2 |
2.2 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 |
1.9 |
GO:0019363 |
pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.2 |
0.6 |
GO:0060368 |
Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.2 |
0.6 |
GO:0052490 |
suppression by virus of host apoptotic process(GO:0019050) negative regulation by symbiont of host apoptotic process(GO:0033668) modulation by virus of host apoptotic process(GO:0039526) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.2 |
0.5 |
GO:0090153 |
regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.2 |
4.5 |
GO:0047496 |
vesicle transport along microtubule(GO:0047496) |
| 0.2 |
1.2 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.2 |
0.6 |
GO:0032804 |
negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) negative regulation of receptor catabolic process(GO:2000645) |
| 0.2 |
2.7 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.2 |
2.6 |
GO:0051205 |
protein insertion into membrane(GO:0051205) |
| 0.2 |
1.5 |
GO:0035774 |
positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.2 |
0.9 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.1 |
1.0 |
GO:0034122 |
negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 |
0.6 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.1 |
3.5 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.1 |
0.3 |
GO:2000273 |
positive regulation of receptor activity(GO:2000273) |
| 0.1 |
0.4 |
GO:1902774 |
late endosome to lysosome transport(GO:1902774) |
| 0.1 |
4.2 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.1 |
2.3 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 |
0.7 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
11.1 |
GO:0071805 |
potassium ion transmembrane transport(GO:0071805) |
| 0.1 |
0.3 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.1 |
0.3 |
GO:1904338 |
regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 |
0.4 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 |
6.7 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.1 |
1.0 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.1 |
1.8 |
GO:0072384 |
organelle transport along microtubule(GO:0072384) |
| 0.1 |
0.8 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 |
1.5 |
GO:0090140 |
regulation of mitochondrial fission(GO:0090140) |
| 0.1 |
0.8 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 |
0.3 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) angiotensin-activated signaling pathway involved in heart process(GO:0086098) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 |
0.4 |
GO:0007620 |
copulation(GO:0007620) |
| 0.1 |
1.3 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.1 |
0.7 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.1 |
0.4 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 |
0.7 |
GO:2000059 |
negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.1 |
0.9 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
0.6 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 |
0.1 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.1 |
2.4 |
GO:0006509 |
membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 |
1.4 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 |
0.8 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
0.2 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.1 |
0.2 |
GO:0016239 |
positive regulation of macroautophagy(GO:0016239) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.1 |
1.9 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 |
1.7 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.1 |
0.5 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
0.5 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.1 |
1.2 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.5 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.1 |
0.4 |
GO:0009227 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.1 |
1.2 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
6.8 |
GO:0014065 |
phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.1 |
0.9 |
GO:0061013 |
regulation of mRNA catabolic process(GO:0061013) positive regulation of mRNA catabolic process(GO:0061014) |
| 0.1 |
0.9 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.1 |
0.1 |
GO:2000546 |
positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 |
0.8 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.1 |
2.4 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.1 |
10.9 |
GO:0046488 |
phosphatidylinositol metabolic process(GO:0046488) |
| 0.1 |
7.4 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.1 |
2.1 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.1 |
1.4 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.1 |
0.4 |
GO:0033004 |
negative regulation of mast cell activation(GO:0033004) |
| 0.1 |
0.6 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.1 |
1.3 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.1 |
0.6 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
1.3 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.1 |
0.8 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
0.4 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.8 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 |
0.6 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 |
0.6 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.1 |
1.4 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 |
1.3 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.1 |
0.9 |
GO:0050995 |
negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 |
2.6 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.1 |
0.2 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 |
0.2 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 |
1.1 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.1 |
0.3 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.1 |
0.5 |
GO:0043649 |
dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 |
0.6 |
GO:0001964 |
startle response(GO:0001964) |
| 0.1 |
2.3 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.1 |
0.2 |
GO:0048703 |
embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.1 |
0.2 |
GO:0090285 |
regulation of protein glycosylation in Golgi(GO:0090283) negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 |
1.6 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.1 |
0.2 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.1 |
1.6 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 |
1.3 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.1 |
0.2 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
0.8 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.1 |
1.1 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.1 |
0.7 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.1 |
0.7 |
GO:2000479 |
regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.1 |
1.3 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 |
0.4 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.3 |
GO:0055012 |
ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.1 |
1.1 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 |
0.5 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 |
0.3 |
GO:0046416 |
D-amino acid metabolic process(GO:0046416) |
| 0.1 |
0.6 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.1 |
0.3 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) histidine transport(GO:0015817) |
| 0.1 |
0.3 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.1 |
0.3 |
GO:0032469 |
endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 |
1.6 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.1 |
0.3 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 |
1.5 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.3 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
0.4 |
GO:0002643 |
tolerance induction(GO:0002507) regulation of tolerance induction(GO:0002643) |
| 0.1 |
0.9 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 |
0.6 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 |
0.2 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 |
0.7 |
GO:0051569 |
regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 |
0.5 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.1 |
0.5 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
0.3 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.9 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.1 |
1.0 |
GO:0060122 |
inner ear receptor stereocilium organization(GO:0060122) |
| 0.1 |
5.1 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.1 |
0.5 |
GO:0006283 |
transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 |
2.8 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 |
0.7 |
GO:0060765 |
regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 |
0.3 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.1 |
1.4 |
GO:0034243 |
regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.1 |
0.1 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.1 |
0.5 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 |
0.7 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.1 |
0.3 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 |
0.5 |
GO:0051974 |
negative regulation of telomerase activity(GO:0051974) |
| 0.1 |
0.3 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.1 |
2.2 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
0.4 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 |
0.5 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.0 |
0.6 |
GO:0043537 |
negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 |
1.6 |
GO:0006497 |
protein lipidation(GO:0006497) |
| 0.0 |
1.2 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 |
0.4 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.3 |
GO:0031440 |
regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
2.4 |
GO:0030705 |
cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 |
0.6 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
5.4 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.3 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.6 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.4 |
GO:0061462 |
protein localization to lysosome(GO:0061462) |
| 0.0 |
0.1 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 |
0.4 |
GO:0016446 |
somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.5 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.3 |
GO:0090280 |
positive regulation of calcium ion import(GO:0090280) |
| 0.0 |
0.3 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.9 |
GO:0048536 |
spleen development(GO:0048536) |
| 0.0 |
0.4 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.5 |
GO:0006744 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 |
0.2 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.4 |
GO:0008206 |
bile acid metabolic process(GO:0008206) |
| 0.0 |
0.2 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.2 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.1 |
GO:0009584 |
detection of visible light(GO:0009584) |
| 0.0 |
0.1 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.1 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 |
0.6 |
GO:0046888 |
negative regulation of hormone secretion(GO:0046888) |
| 0.0 |
0.1 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 |
0.4 |
GO:0007018 |
microtubule-based movement(GO:0007018) |
| 0.0 |
0.2 |
GO:0015914 |
organophosphate ester transport(GO:0015748) phospholipid transport(GO:0015914) |
| 0.0 |
0.1 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.1 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.0 |
GO:0030220 |
platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 |
0.1 |
GO:0051642 |
centrosome localization(GO:0051642) |
| 0.0 |
0.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.1 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.1 |
GO:2000637 |
positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 |
0.0 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |