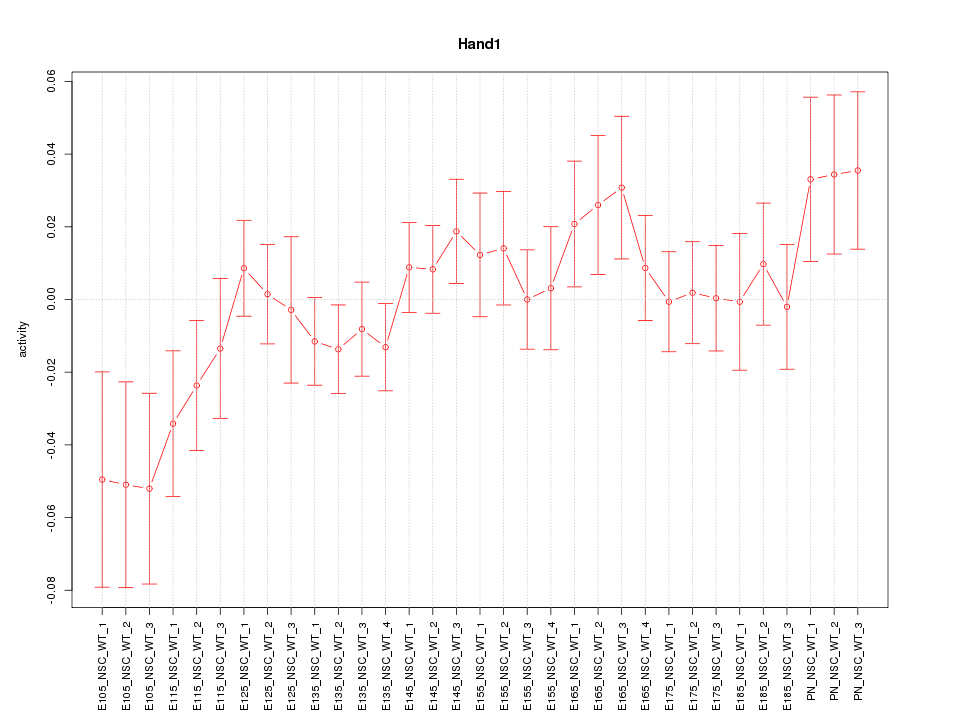
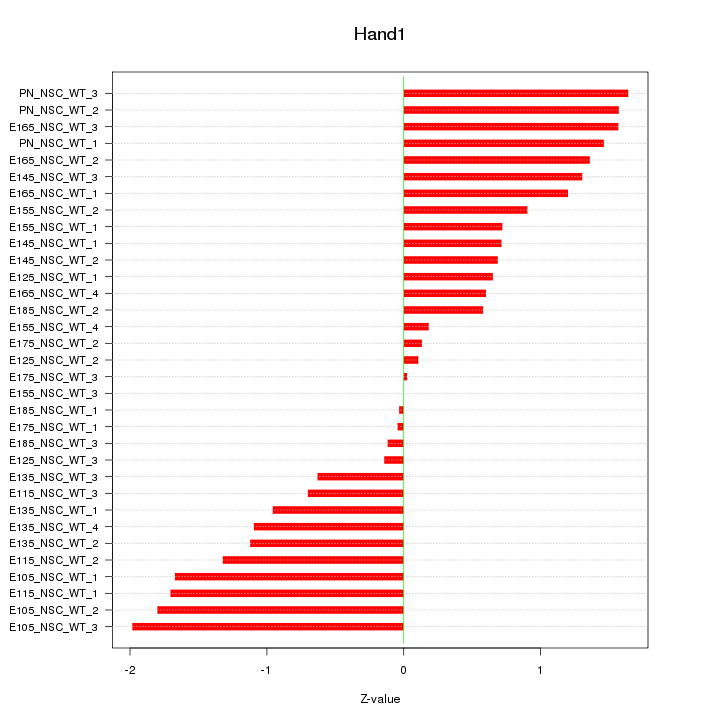
Motif ID: Hand1
Z-value: 1.062
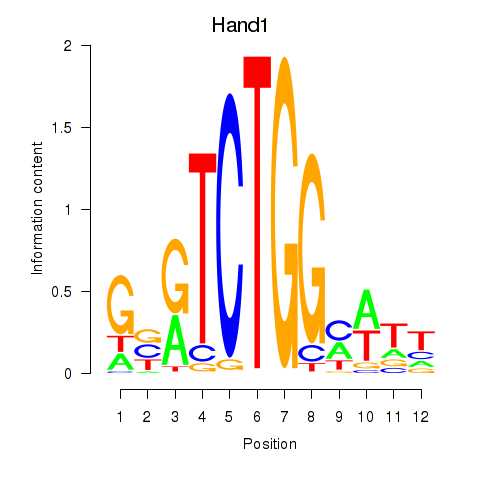
Transcription factors associated with Hand1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hand1 | ENSMUSG00000037335.7 | Hand1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 1.4 | 4.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 1.2 | 4.9 | GO:0042938 | dipeptide transport(GO:0042938) |
| 1.1 | 4.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.1 | 3.3 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 1.1 | 3.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.0 | 3.9 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.9 | 5.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.9 | 3.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.8 | 6.7 | GO:2000809 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.8 | 2.3 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.8 | 2.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.7 | 5.2 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.7 | 2.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.7 | 2.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.7 | 2.0 | GO:0071544 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.6 | 3.8 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.6 | 1.8 | GO:0060023 | soft palate development(GO:0060023) |
| 0.6 | 2.3 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.6 | 1.7 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.5 | 4.2 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.5 | 3.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.4 | 3.5 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.4 | 1.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 3.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 4.0 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.4 | 2.3 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.4 | 1.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 2.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.3 | 5.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 1.4 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.3 | 7.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 1.1 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.3 | 6.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.3 | 1.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.3 | 4.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.3 | 1.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.3 | 1.5 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.2 | 1.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 1.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 1.8 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 | 0.7 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 0.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.9 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.2 | 0.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.2 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 5.4 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.2 | 1.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.7 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.2 | 4.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 4.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 2.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.2 | 1.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 3.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.3 | GO:1900426 | positive regulation of defense response to bacterium(GO:1900426) |
| 0.1 | 1.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 13.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 2.2 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.1 | 0.8 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.4 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.4 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 1.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 1.9 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 1.6 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.3 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.9 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 1.6 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.1 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.1 | 0.4 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.1 | 0.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 3.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 2.6 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 1.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.7 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.6 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 0.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.4 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glycolytic process through glucose-1-phosphate(GO:0061622) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) actin filament severing(GO:0051014) |
| 0.1 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.3 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 1.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 2.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 2.5 | GO:0090003 | regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 1.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 2.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.1 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 2.0 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 3.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 4.1 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.4 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.7 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.8 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 10.6 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.3 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 1.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 2.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 3.8 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.5 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.2 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 1.9 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 1.3 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 1.4 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.3 | 3.8 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 1.2 | 7.2 | GO:0044308 | axonal spine(GO:0044308) |
| 1.2 | 3.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.7 | 2.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.6 | 3.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 4.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 1.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.4 | 2.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 4.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 5.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 1.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.3 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 2.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 4.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 2.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 4.9 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 1.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 2.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.3 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 5.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 2.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 3.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 4.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 3.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 1.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 7.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 3.6 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 4.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 3.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 1.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 5.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 4.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.1 | 6.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.1 | 3.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.0 | 4.9 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.9 | 3.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.8 | 5.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.8 | 2.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.7 | 4.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.6 | 2.4 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.6 | 1.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.5 | 1.6 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.5 | 3.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.5 | 3.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 4.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.5 | 1.8 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.4 | 3.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.4 | 1.9 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.4 | 1.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 2.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 2.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.3 | 5.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 1.9 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 1.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.8 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 1.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 3.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.4 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.2 | 0.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 2.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 4.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 3.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 3.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 2.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 2.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 3.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 5.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 2.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.6 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.6 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 2.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.4 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 4.7 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 4.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 8.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 5.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 5.6 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.8 | GO:0017161 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) inositol-4,5-bisphosphate 5-phosphatase activity(GO:0030487) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 1.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0050308 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 2.1 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 2.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |