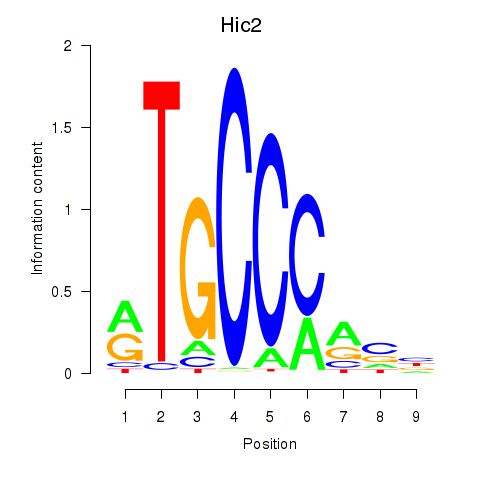
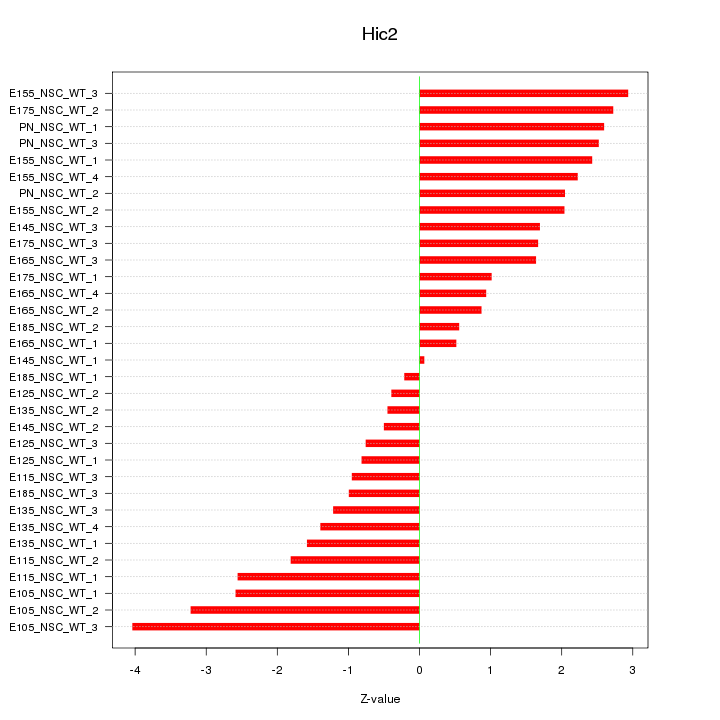
| 4.3 |
13.0 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 3.8 |
15.2 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 3.2 |
3.2 |
GO:0010593 |
negative regulation of lamellipodium assembly(GO:0010593) |
| 3.0 |
48.6 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 2.1 |
2.1 |
GO:1900020 |
regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 2.1 |
6.2 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.8 |
7.4 |
GO:0021586 |
pons maturation(GO:0021586) |
| 1.8 |
9.0 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.8 |
5.3 |
GO:1903054 |
negative regulation of extracellular matrix organization(GO:1903054) |
| 1.7 |
6.9 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 1.6 |
6.6 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 1.6 |
1.6 |
GO:0051926 |
negative regulation of calcium ion transport(GO:0051926) |
| 1.5 |
9.1 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 1.5 |
5.8 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 1.4 |
2.9 |
GO:0035990 |
tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 1.4 |
8.4 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 1.4 |
6.8 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 1.3 |
4.0 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 1.3 |
5.3 |
GO:2001106 |
regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.3 |
3.9 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 1.3 |
12.6 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.2 |
3.7 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.2 |
3.6 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 1.2 |
6.0 |
GO:1900242 |
regulation of synaptic vesicle endocytosis(GO:1900242) |
| 1.1 |
3.4 |
GO:0006116 |
NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.1 |
3.4 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 1.1 |
4.4 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 1.1 |
3.2 |
GO:2000344 |
positive regulation of acrosome reaction(GO:2000344) |
| 1.1 |
3.2 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 1.0 |
3.1 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.0 |
9.2 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 1.0 |
15.1 |
GO:0021957 |
corticospinal tract morphogenesis(GO:0021957) |
| 1.0 |
6.8 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.0 |
2.9 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 1.0 |
2.9 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.9 |
7.5 |
GO:0071435 |
potassium ion export(GO:0071435) |
| 0.9 |
3.7 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.9 |
3.7 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.9 |
7.3 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.9 |
2.7 |
GO:0034334 |
adherens junction maintenance(GO:0034334) |
| 0.9 |
4.5 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.9 |
4.5 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.9 |
2.7 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.9 |
8.7 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.9 |
2.6 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.8 |
3.3 |
GO:0071072 |
negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.8 |
3.2 |
GO:1903802 |
positive regulation of arachidonic acid secretion(GO:0090238) L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.8 |
12.1 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.8 |
1.5 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.8 |
2.3 |
GO:0046544 |
development of secondary male sexual characteristics(GO:0046544) |
| 0.8 |
1.5 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.7 |
3.0 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.7 |
2.2 |
GO:1902083 |
negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.7 |
3.6 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.7 |
2.9 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.7 |
4.2 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.7 |
2.1 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.7 |
2.8 |
GO:0045188 |
circadian sleep/wake cycle, non-REM sleep(GO:0042748) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.7 |
4.2 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.7 |
5.5 |
GO:0032261 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.7 |
2.0 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.7 |
0.7 |
GO:0050912 |
detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.7 |
2.0 |
GO:1903061 |
regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.7 |
2.0 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.7 |
2.0 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.7 |
3.3 |
GO:1903598 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) regulation of inward rectifier potassium channel activity(GO:1901979) positive regulation of gap junction assembly(GO:1903598) |
| 0.6 |
1.3 |
GO:1902995 |
regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.6 |
10.4 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.6 |
1.8 |
GO:0051542 |
elastin biosynthetic process(GO:0051542) |
| 0.6 |
5.5 |
GO:2000489 |
hepatic stellate cell activation(GO:0035733) regulation of hepatic stellate cell activation(GO:2000489) |
| 0.6 |
3.0 |
GO:0034372 |
very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.6 |
3.0 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.6 |
3.6 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.6 |
2.4 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.6 |
0.6 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.6 |
4.1 |
GO:0038171 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.6 |
1.8 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.6 |
2.3 |
GO:0050913 |
sensory perception of bitter taste(GO:0050913) |
| 0.6 |
1.7 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.6 |
5.6 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.6 |
3.3 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.5 |
3.8 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.5 |
2.2 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.5 |
3.8 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.5 |
4.2 |
GO:0045187 |
regulation of circadian sleep/wake cycle, sleep(GO:0045187) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.5 |
1.5 |
GO:0090403 |
oxidative stress-induced premature senescence(GO:0090403) |
| 0.5 |
1.0 |
GO:1990705 |
cholangiocyte proliferation(GO:1990705) |
| 0.5 |
1.5 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.5 |
0.5 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.5 |
1.4 |
GO:0010835 |
regulation of protein ADP-ribosylation(GO:0010835) |
| 0.5 |
4.3 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.5 |
1.4 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.5 |
1.9 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.5 |
1.9 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.5 |
1.9 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 |
3.7 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.5 |
5.1 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.5 |
1.9 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.5 |
2.3 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) taurine metabolic process(GO:0019530) response to glucagon(GO:0033762) |
| 0.4 |
3.1 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.4 |
1.3 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.4 |
10.2 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 |
2.2 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.4 |
2.2 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.4 |
8.6 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.4 |
2.1 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.4 |
1.3 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 |
1.7 |
GO:0034727 |
lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.4 |
1.2 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.4 |
0.8 |
GO:1900451 |
positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.4 |
2.9 |
GO:1900452 |
regulation of long term synaptic depression(GO:1900452) |
| 0.4 |
2.8 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 |
2.8 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.4 |
2.4 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.4 |
1.5 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.4 |
1.5 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.4 |
1.5 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.4 |
3.4 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.4 |
0.4 |
GO:0050883 |
musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.4 |
1.9 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.4 |
1.5 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.4 |
2.2 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.4 |
1.5 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.4 |
0.7 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.4 |
0.7 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.4 |
1.4 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.4 |
1.8 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.3 |
1.4 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.3 |
1.4 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.3 |
3.7 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.3 |
3.0 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.3 |
1.0 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.3 |
3.9 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.3 |
0.7 |
GO:0070100 |
negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.3 |
2.3 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.3 |
1.0 |
GO:0060785 |
regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.3 |
4.8 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.3 |
0.9 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.3 |
4.6 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.3 |
2.1 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.3 |
1.5 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.3 |
1.2 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.3 |
10.5 |
GO:0035640 |
exploration behavior(GO:0035640) |
| 0.3 |
1.8 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 |
2.4 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.3 |
1.2 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.3 |
2.6 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.3 |
0.9 |
GO:1901509 |
branch elongation involved in ureteric bud branching(GO:0060681) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 |
2.9 |
GO:0021940 |
positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.3 |
0.9 |
GO:0035435 |
phosphate ion transmembrane transport(GO:0035435) |
| 0.3 |
2.0 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 |
1.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 |
5.6 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.3 |
2.8 |
GO:0007274 |
neuromuscular synaptic transmission(GO:0007274) |
| 0.3 |
0.8 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 |
2.7 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.3 |
3.6 |
GO:0033198 |
response to ATP(GO:0033198) |
| 0.3 |
1.5 |
GO:0046103 |
ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.3 |
0.5 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.3 |
5.1 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 |
1.0 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.2 |
4.9 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.2 |
2.5 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 |
0.7 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 |
1.9 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 |
1.8 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.2 |
5.3 |
GO:0034113 |
heterotypic cell-cell adhesion(GO:0034113) |
| 0.2 |
1.1 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.2 |
2.0 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.2 |
1.6 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.2 |
3.0 |
GO:0032438 |
melanosome organization(GO:0032438) |
| 0.2 |
1.1 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.2 |
3.4 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.2 |
1.3 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.2 |
2.9 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 |
1.4 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.2 |
2.6 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 |
2.8 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.2 |
2.5 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.2 |
1.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.2 |
0.4 |
GO:0061043 |
regulation of vascular wound healing(GO:0061043) |
| 0.2 |
3.6 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.2 |
2.3 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 |
2.6 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 |
2.1 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.2 |
6.3 |
GO:1901381 |
positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.2 |
5.0 |
GO:0060749 |
mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.2 |
1.1 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 |
0.2 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.2 |
0.5 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.2 |
3.3 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 |
2.9 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 |
1.9 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 |
1.0 |
GO:1900273 |
positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.2 |
0.3 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) |
| 0.2 |
4.8 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.2 |
0.8 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 |
3.5 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.2 |
0.5 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 |
6.7 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.2 |
6.0 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.2 |
0.5 |
GO:1901386 |
negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.2 |
2.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.2 |
0.9 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.2 |
0.8 |
GO:1903772 |
regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.2 |
0.3 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 |
2.4 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.1 |
0.3 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
6.0 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.1 |
0.6 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.1 |
1.5 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
0.9 |
GO:0044829 |
positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 |
5.1 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.1 |
0.4 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
2.3 |
GO:0006024 |
glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 |
4.6 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.1 |
1.3 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.1 |
4.8 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.1 |
0.7 |
GO:0019086 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.1 |
1.5 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.1 |
1.4 |
GO:0060294 |
cilium movement involved in cell motility(GO:0060294) |
| 0.1 |
1.6 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
3.6 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.1 |
0.5 |
GO:0071442 |
regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 |
1.1 |
GO:0046928 |
regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 |
0.7 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.1 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 |
0.1 |
GO:1905154 |
negative regulation of membrane invagination(GO:1905154) |
| 0.1 |
1.2 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 |
0.9 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 |
0.5 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.1 |
1.5 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 |
0.4 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.1 |
2.0 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 |
0.7 |
GO:0001880 |
Mullerian duct regression(GO:0001880) |
| 0.1 |
1.8 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.1 |
0.5 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.1 |
0.8 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 |
0.6 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 |
0.7 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
0.8 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 |
0.8 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
1.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 |
0.8 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 |
0.8 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 |
1.1 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 |
1.3 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.1 |
0.3 |
GO:1902172 |
keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 |
5.1 |
GO:0001541 |
ovarian follicle development(GO:0001541) |
| 0.1 |
1.1 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 |
0.8 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.3 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 |
0.3 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.1 |
0.6 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 |
1.9 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.1 |
0.4 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 |
0.3 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 |
2.8 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.1 |
0.6 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 |
0.8 |
GO:0060836 |
lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 |
0.5 |
GO:0050716 |
positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 |
0.2 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
2.0 |
GO:0010955 |
negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.1 |
1.4 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.1 |
1.1 |
GO:0036065 |
fucosylation(GO:0036065) |
| 0.1 |
0.3 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.1 |
0.8 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.1 |
0.6 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.1 |
0.4 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.6 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
0.4 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
2.4 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 |
0.6 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 |
2.2 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
4.1 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.1 |
0.2 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.1 |
0.9 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.1 |
1.0 |
GO:0042220 |
response to cocaine(GO:0042220) |
| 0.1 |
0.7 |
GO:0032801 |
receptor catabolic process(GO:0032801) |
| 0.1 |
0.8 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
1.2 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.1 |
1.1 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.1 |
1.0 |
GO:0001658 |
branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.1 |
0.9 |
GO:0014898 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 |
1.0 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.2 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
1.7 |
GO:0051220 |
cytoplasmic sequestering of protein(GO:0051220) |
| 0.1 |
0.7 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.1 |
0.7 |
GO:0090218 |
positive regulation of lipid kinase activity(GO:0090218) |
| 0.1 |
0.7 |
GO:0003094 |
glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.1 |
0.7 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 |
1.8 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 |
2.0 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.1 |
2.0 |
GO:0006509 |
membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 |
0.3 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.1 |
1.7 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.1 |
0.2 |
GO:0035459 |
cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
1.1 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.9 |
GO:0042832 |
defense response to protozoan(GO:0042832) |
| 0.0 |
0.2 |
GO:0071267 |
amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 |
0.3 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.3 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.0 |
0.6 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.2 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
1.1 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.0 |
0.2 |
GO:2001223 |
negative regulation of neuron migration(GO:2001223) |
| 0.0 |
1.8 |
GO:0030593 |
neutrophil chemotaxis(GO:0030593) |
| 0.0 |
0.6 |
GO:0032469 |
endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 |
0.9 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.9 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.3 |
GO:0046716 |
muscle cell cellular homeostasis(GO:0046716) |
| 0.0 |
0.1 |
GO:0008655 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 |
1.7 |
GO:0045839 |
negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 |
0.5 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.3 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.4 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 |
0.1 |
GO:0039535 |
regulation of RIG-I signaling pathway(GO:0039535) |
| 0.0 |
1.4 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.1 |
GO:1903432 |
TORC1 signaling(GO:0038202) regulation of TORC1 signaling(GO:1903432) positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 |
1.0 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
5.4 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.2 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.6 |
GO:0032402 |
establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) |
| 0.0 |
1.2 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.4 |
GO:0021781 |
glial cell fate commitment(GO:0021781) |
| 0.0 |
0.5 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.2 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 |
0.1 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 |
0.1 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.5 |
GO:0019884 |
antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 |
0.6 |
GO:0031663 |
lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 |
1.3 |
GO:0055072 |
iron ion homeostasis(GO:0055072) |
| 0.0 |
0.2 |
GO:0030174 |
regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 |
0.8 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 |
0.1 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 |
0.6 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
1.1 |
GO:0006497 |
protein lipidation(GO:0006497) |
| 0.0 |
0.1 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 |
0.2 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.9 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.1 |
GO:0060821 |
inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.0 |
0.2 |
GO:1901201 |
regulation of extracellular matrix assembly(GO:1901201) |
| 0.0 |
0.4 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
0.3 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.3 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.0 |
0.1 |
GO:0034312 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 |
0.1 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.0 |
0.1 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.0 |
0.6 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.0 |
0.4 |
GO:0006911 |
phagocytosis, engulfment(GO:0006911) |
| 0.0 |
0.3 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 |
0.1 |
GO:0070528 |
protein kinase C signaling(GO:0070528) |
| 0.0 |
0.1 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.0 |
2.2 |
GO:0010976 |
positive regulation of neuron projection development(GO:0010976) |
| 0.0 |
0.1 |
GO:0014068 |
positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 |
1.1 |
GO:0034284 |
response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 |
0.0 |
GO:0051295 |
cleavage furrow formation(GO:0036089) establishment of meiotic spindle localization(GO:0051295) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 |
0.1 |
GO:0042126 |
nitrate metabolic process(GO:0042126) |
| 0.0 |
0.1 |
GO:0010667 |
negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 |
0.4 |
GO:0042733 |
embryonic digit morphogenesis(GO:0042733) |
| 0.0 |
0.1 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.4 |
GO:0032755 |
positive regulation of interleukin-6 production(GO:0032755) |