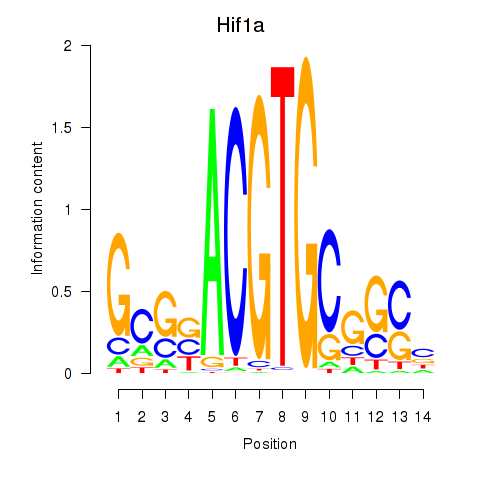
Motif ID: Hif1a
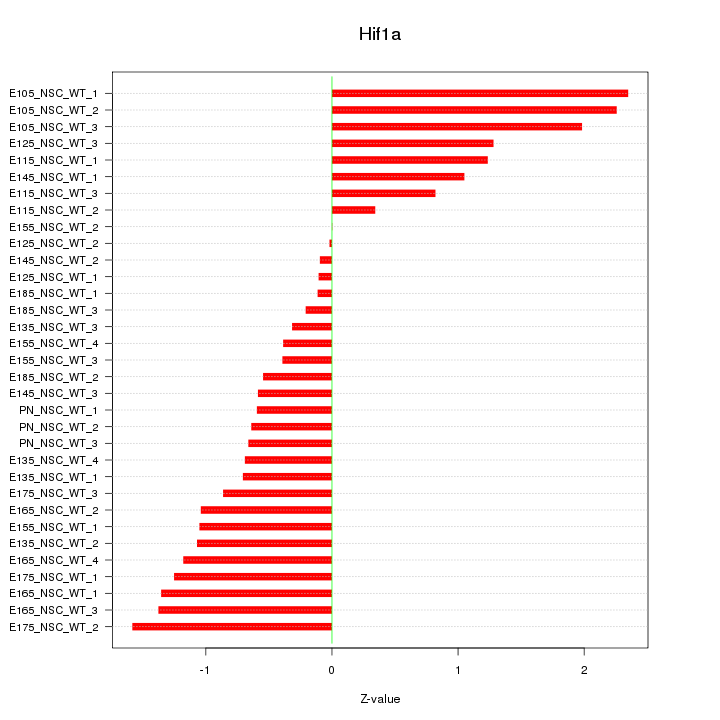
Z-value: 1.047
Transcription factors associated with Hif1a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hif1a | ENSMUSG00000021109.7 | Hif1a |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hif1a | mm10_v2_chr12_+_73907904_73907970 | -0.84 | 8.5e-10 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 2.0 | 8.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.9 | 7.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.8 | 9.1 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.8 | 7.1 | GO:0032439 | endosome localization(GO:0032439) |
| 1.6 | 1.6 | GO:0033122 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 1.6 | 4.8 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 1.6 | 4.7 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 1.5 | 4.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.2 | 3.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.0 | 3.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.9 | 2.7 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.9 | 1.8 | GO:0002842 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.6 | 1.9 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.6 | 3.0 | GO:0042117 | hyaluronan catabolic process(GO:0030214) monocyte activation(GO:0042117) |
| 0.6 | 1.8 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.6 | 4.6 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.5 | 1.0 | GO:2000224 | testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.5 | 1.5 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.5 | 2.0 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.5 | 1.5 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) response to cobalt ion(GO:0032025) |
| 0.5 | 1.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.5 | 1.4 | GO:0071336 | lung growth(GO:0060437) positive regulation of fat cell proliferation(GO:0070346) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.4 | 2.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.4 | 3.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 4.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.4 | 1.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.4 | 2.4 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.4 | 3.8 | GO:0030388 | fructose 6-phosphate metabolic process(GO:0006002) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 2.6 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) negative regulation of monocyte differentiation(GO:0045656) negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.4 | 2.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.4 | 1.8 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 1.4 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.3 | 3.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.3 | 1.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 3.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.3 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.3 | 1.6 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.3 | 18.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.3 | 0.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of cellular pH reduction(GO:0032847) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 2.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.3 | 2.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.3 | 2.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 2.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 0.6 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 3.5 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.2 | 0.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 0.4 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.2 | 0.7 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 2.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.2 | 3.8 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.2 | 0.5 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 0.7 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 1.0 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.2 | 5.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.2 | 0.8 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.2 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 0.9 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 2.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 4.7 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.4 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 2.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 5.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.5 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.1 | 1.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.5 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 1.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 1.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 1.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 5.8 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 1.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.3 | GO:0061198 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) fungiform papilla formation(GO:0061198) |
| 0.1 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 1.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 1.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 7.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 1.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 0.4 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.1 | 7.3 | GO:0071229 | cellular response to acid chemical(GO:0071229) |
| 0.0 | 0.5 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 1.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 5.1 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 1.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.4 | GO:1903513 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 1.3 | GO:0010470 | regulation of gastrulation(GO:0010470) |
| 0.0 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 2.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 1.0 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.8 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.5 | 4.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.4 | 4.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 1.2 | 7.1 | GO:0045179 | apical cortex(GO:0045179) |
| 1.1 | 3.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.1 | 7.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.0 | 6.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.9 | 3.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.7 | 3.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.7 | 3.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.7 | 2.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.6 | 9.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.6 | 4.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.5 | 4.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.4 | 1.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 2.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 3.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 2.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 1.9 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 2.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.0 | GO:0048188 | MLL3/4 complex(GO:0044666) Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 1.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 2.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 3.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 4.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 3.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 2.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 2.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 5.5 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 4.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 21.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.1 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 1.5 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 1.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.5 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.5 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.8 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 2.5 | 7.5 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 2.3 | 9.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 2.0 | 8.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.6 | 4.7 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.9 | 3.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.9 | 3.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.8 | 7.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.8 | 3.0 | GO:0030292 | hyalurononglucosaminidase activity(GO:0004415) protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.6 | 3.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.6 | 10.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.5 | 1.6 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.5 | 2.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 2.2 | GO:0070883 | importin-alpha family protein binding(GO:0061676) pre-miRNA binding(GO:0070883) |
| 0.4 | 4.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 1.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 1.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 1.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.3 | 3.1 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.3 | 1.9 | GO:0002135 | CTP binding(GO:0002135) |
| 0.3 | 4.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 3.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 1.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.3 | 1.0 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 2.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.6 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.2 | 1.8 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 2.0 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 0.9 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.2 | 2.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 3.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 6.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 2.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.2 | 1.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 2.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.7 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 0.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 2.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 7.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 4.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.6 | GO:0017161 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.1 | 1.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 4.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 6.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.7 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.1 | 0.6 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.1 | 1.7 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 1.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 7.5 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.3 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 2.1 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.8 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 2.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.9 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 1.4 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 5.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |