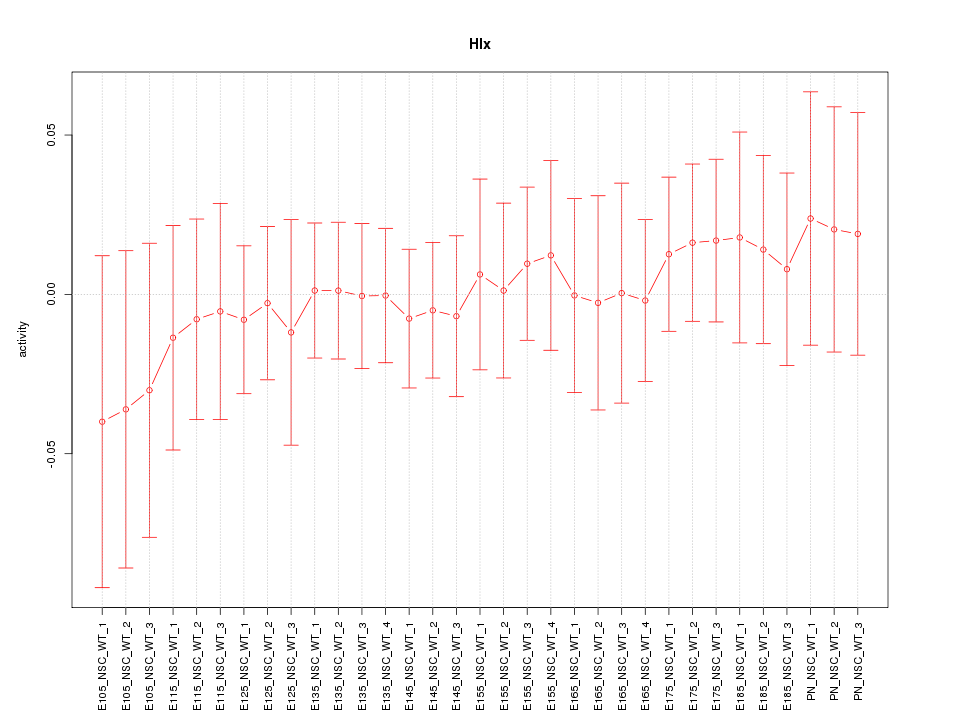
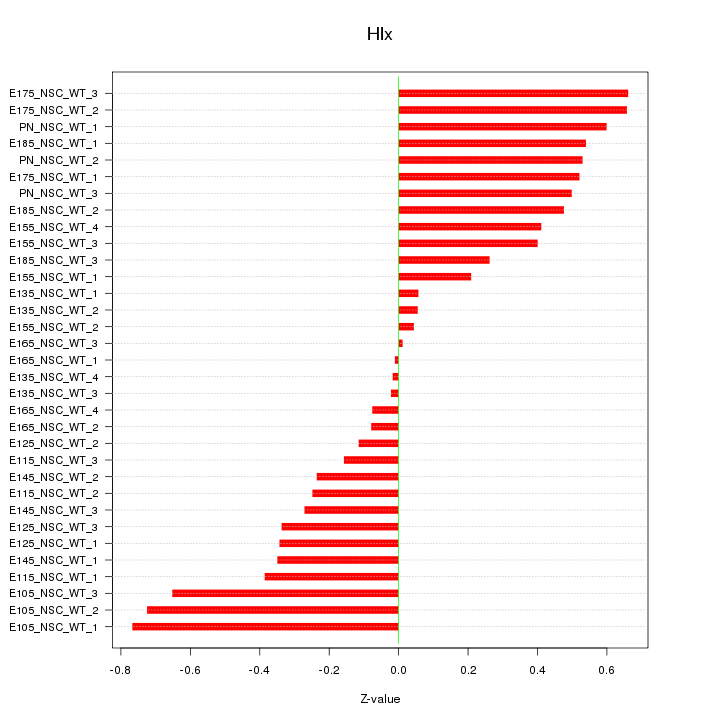
Motif ID: Hlx
Z-value: 0.400

Transcription factors associated with Hlx:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hlx | ENSMUSG00000039377.6 | Hlx |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hlx | mm10_v2_chr1_-_184732616_184732627 | 0.50 | 2.8e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.3 | 5.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 0.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 0.5 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.4 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 1.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.5 | GO:0043205 | fibril(GO:0043205) |
| 0.2 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 1.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 5.4 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 5.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 1.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |