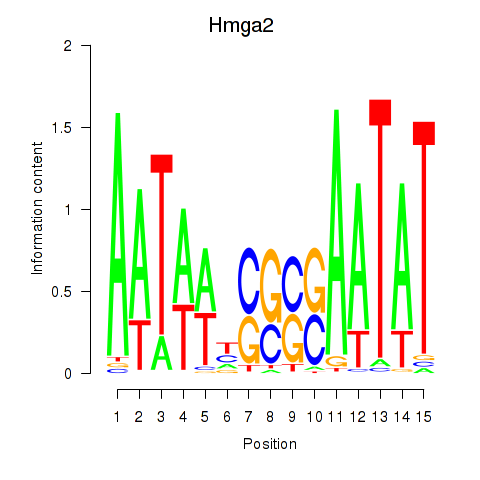
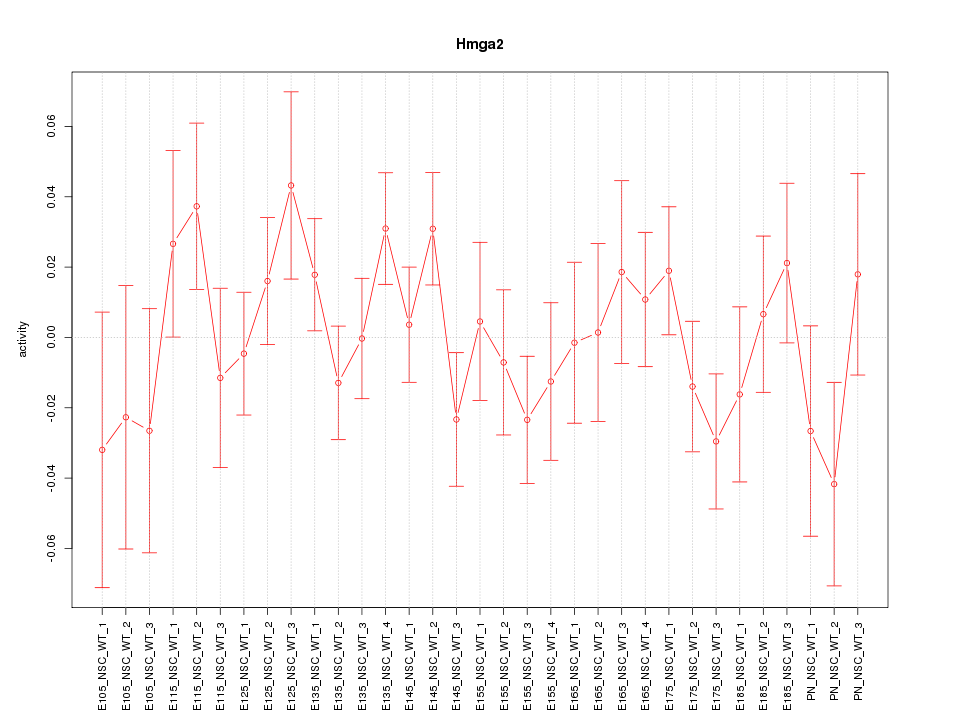
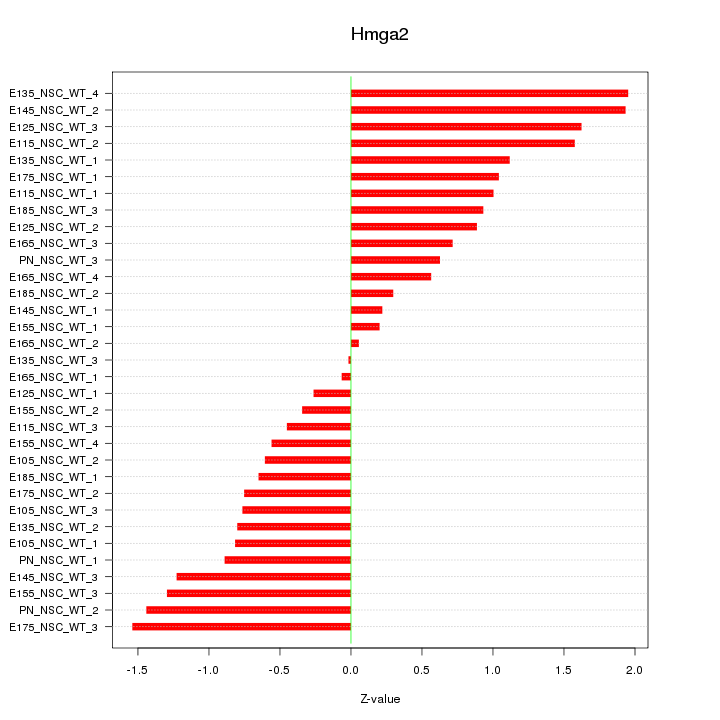
| 0.3 |
0.9 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.3 |
1.1 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.2 |
1.2 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.2 |
1.0 |
GO:0010519 |
negative regulation of phospholipase activity(GO:0010519) |
| 0.2 |
0.9 |
GO:0032929 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.4 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) positive regulation of cortisol secretion(GO:0051464) |
| 0.1 |
0.5 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
0.4 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 |
0.9 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.1 |
0.3 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.1 |
0.8 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 |
0.9 |
GO:0010807 |
regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 |
0.3 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.1 |
0.6 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 |
0.3 |
GO:2000427 |
regulation of apoptotic cell clearance(GO:2000425) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
0.2 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 |
0.2 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.0 |
0.2 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.0 |
0.1 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.0 |
0.7 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.0 |
0.4 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
0.5 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.3 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.4 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.8 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
0.5 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) cGMP catabolic process(GO:0046069) |
| 0.0 |
0.1 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 |
0.3 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.0 |
0.3 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.1 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.0 |
0.1 |
GO:0017121 |
phospholipid scrambling(GO:0017121) |
| 0.0 |
0.4 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.5 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.6 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.5 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.4 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.4 |
GO:0034605 |
cellular response to heat(GO:0034605) |