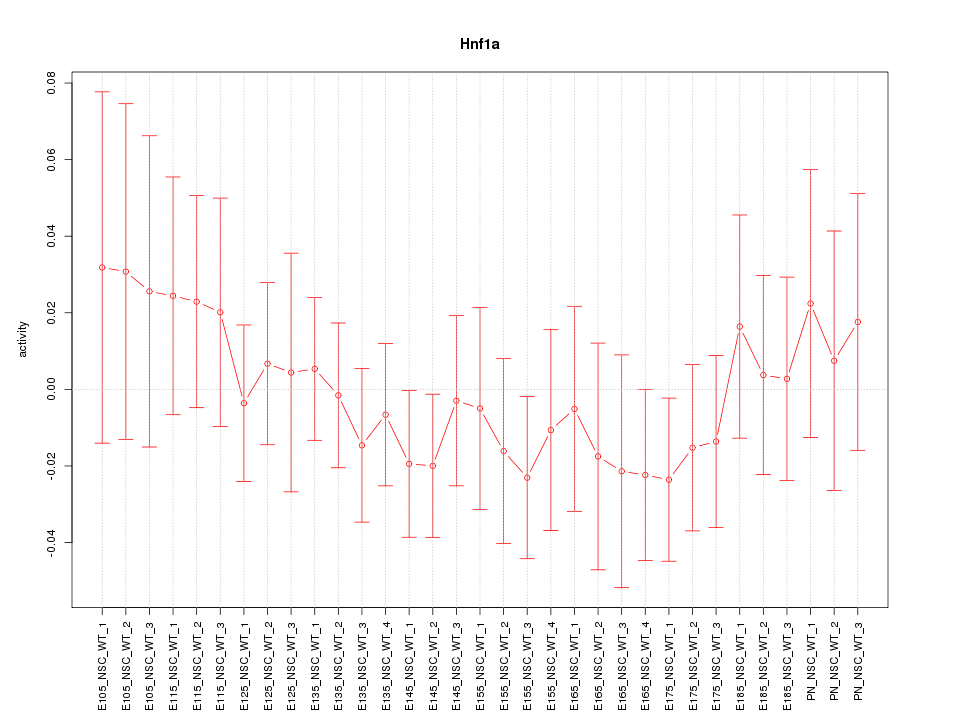
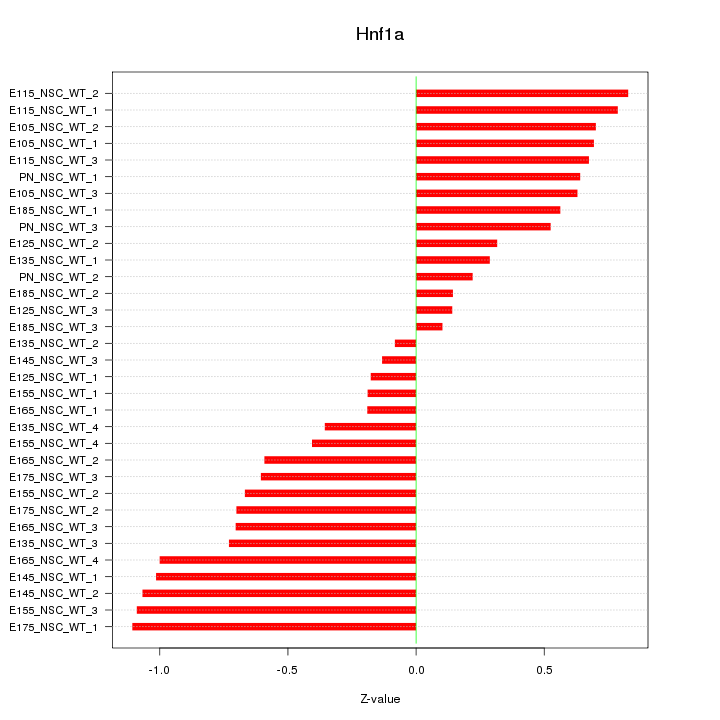
Motif ID: Hnf1a
Z-value: 0.630
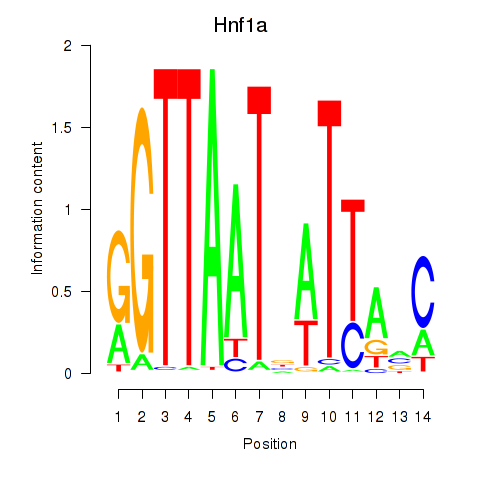
Transcription factors associated with Hnf1a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hnf1a | ENSMUSG00000029556.6 | Hnf1a |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.8 | 3.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.7 | 2.9 | GO:1904192 | prostate gland stromal morphogenesis(GO:0060741) cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.5 | 1.6 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.4 | 2.0 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 2.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 2.0 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 0.7 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 0.8 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.2 | 0.6 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.2 | 1.3 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.2 | 0.6 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.2 | 2.0 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.2 | 1.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.4 | GO:0003195 | tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.7 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.8 | GO:0032782 | urea transport(GO:0015840) bile acid secretion(GO:0032782) urea transmembrane transport(GO:0071918) |
| 0.1 | 0.9 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) negative regulation of superoxide anion generation(GO:0032929) negative regulation of vasodilation(GO:0045908) |
| 0.0 | 1.4 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.5 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.1 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 | 1.0 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.8 | GO:0006821 | chloride transport(GO:0006821) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.7 | 2.0 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.4 | 1.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 3.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 3.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.7 | 2.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.5 | 2.0 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.4 | 2.7 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 0.8 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.4 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.8 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 2.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.5 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 2.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 5.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 2.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.1 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 1.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 1.4 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |