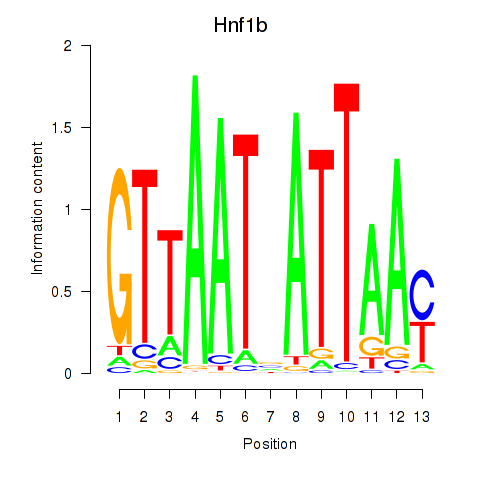
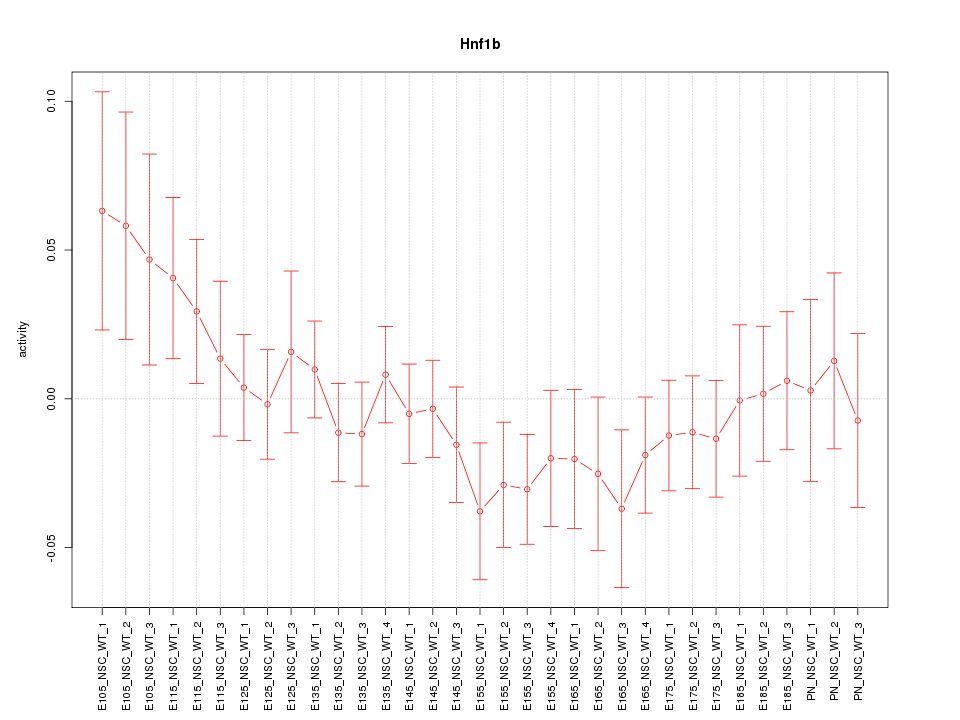
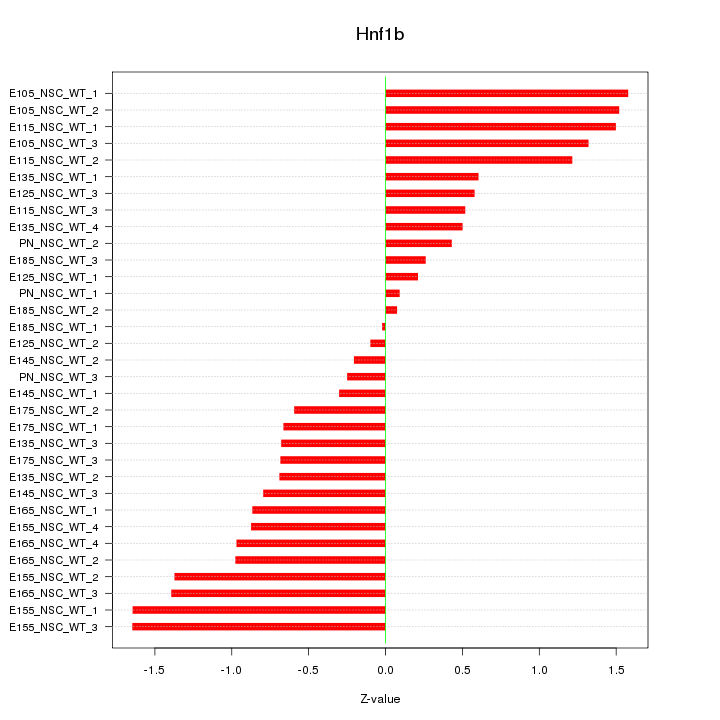
| 1.7 |
5.0 |
GO:1902396 |
protein localization to bicellular tight junction(GO:1902396) |
| 1.6 |
4.8 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 1.5 |
4.5 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.0 |
2.9 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.0 |
2.9 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.9 |
4.4 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.9 |
4.3 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.7 |
2.1 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.5 |
2.7 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 |
2.6 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.5 |
1.6 |
GO:0050787 |
glycolate metabolic process(GO:0009441) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) glyoxal catabolic process(GO:1903190) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) positive regulation of androgen receptor activity(GO:2000825) |
| 0.5 |
1.5 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.5 |
2.9 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.4 |
1.3 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.4 |
1.1 |
GO:1902369 |
negative regulation of RNA catabolic process(GO:1902369) |
| 0.3 |
1.7 |
GO:0072757 |
cellular response to camptothecin(GO:0072757) |
| 0.3 |
4.0 |
GO:0034383 |
low-density lipoprotein particle clearance(GO:0034383) |
| 0.3 |
2.3 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 |
1.2 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 |
3.3 |
GO:0042178 |
xenobiotic catabolic process(GO:0042178) |
| 0.2 |
1.3 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.2 |
1.3 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.2 |
1.3 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 |
2.1 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 |
1.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.2 |
1.4 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.2 |
1.0 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.2 |
0.5 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.1 |
4.0 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 |
1.3 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.1 |
2.5 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
1.1 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.1 |
1.1 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 |
0.5 |
GO:0044830 |
modulation by host of viral RNA genome replication(GO:0044830) |
| 0.1 |
0.7 |
GO:0052695 |
cellular glucuronidation(GO:0052695) |
| 0.1 |
0.7 |
GO:0046689 |
response to mercury ion(GO:0046689) |
| 0.1 |
1.0 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.1 |
0.9 |
GO:0033147 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 |
1.1 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.1 |
0.3 |
GO:0050748 |
negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.1 |
0.4 |
GO:0036112 |
medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 |
0.6 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
0.5 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
2.8 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
1.4 |
GO:0072698 |
protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 |
0.8 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.2 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
0.2 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 |
1.1 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.0 |
0.5 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
2.2 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.2 |
GO:0034472 |
snRNA 3'-end processing(GO:0034472) |
| 0.0 |
0.4 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.1 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.0 |
0.5 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.1 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 |
0.4 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
1.2 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 |
0.4 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.4 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.1 |
GO:1904956 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) regulation of endodermal cell fate specification(GO:0042663) motor learning(GO:0061743) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 |
0.1 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 |
1.0 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
2.4 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.1 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
1.5 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.0 |
0.0 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 |
0.1 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.0 |
0.1 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.0 |
2.8 |
GO:0042384 |
cilium assembly(GO:0042384) |
| 0.0 |
0.2 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |