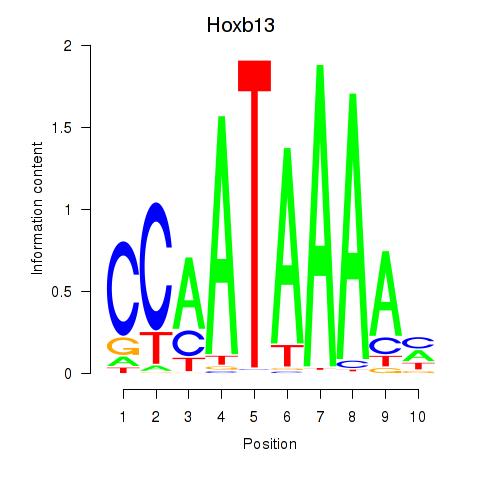
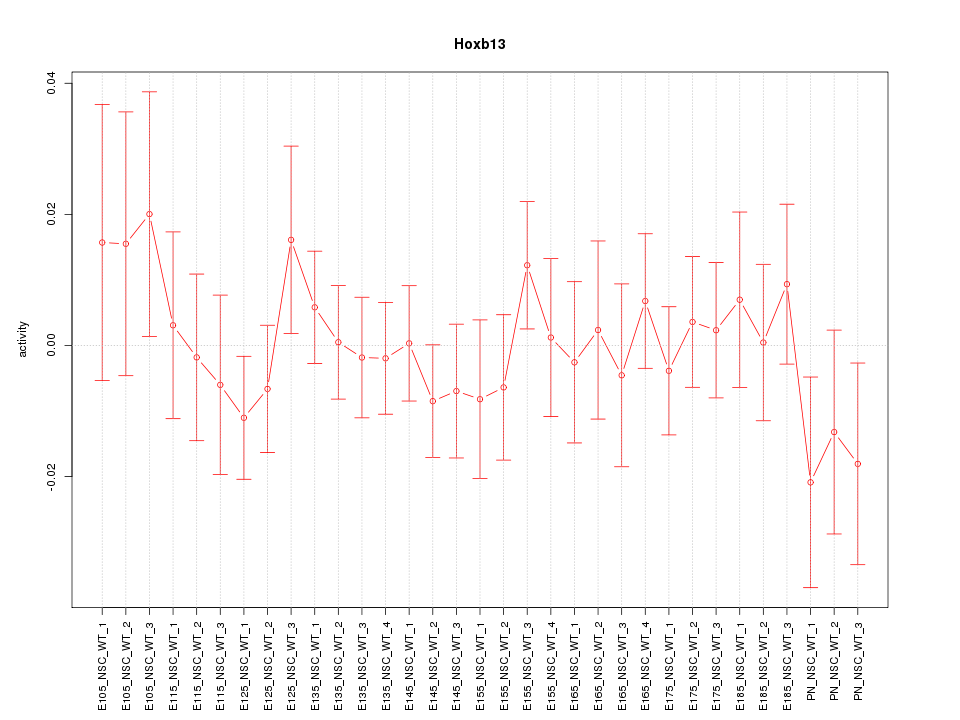
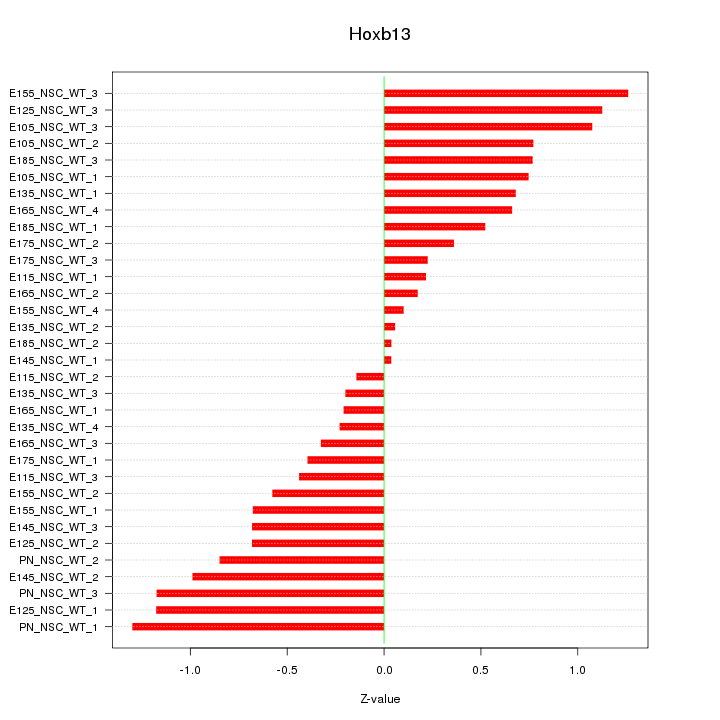
| 0.8 |
2.4 |
GO:0060769 |
positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.6 |
1.7 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.6 |
3.3 |
GO:2000275 |
regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.5 |
1.9 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.4 |
0.8 |
GO:1902956 |
regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.4 |
1.4 |
GO:0006529 |
asparagine biosynthetic process(GO:0006529) |
| 0.3 |
1.0 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 |
1.6 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.3 |
1.0 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.3 |
1.2 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 |
0.9 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 |
0.9 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) |
| 0.3 |
1.7 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.3 |
1.4 |
GO:0046864 |
retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.3 |
1.1 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.3 |
1.1 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.3 |
0.8 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.2 |
0.7 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 |
0.7 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 |
0.7 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.2 |
0.6 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.2 |
0.6 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.2 |
2.4 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.2 |
0.9 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.2 |
0.7 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 |
0.5 |
GO:0048698 |
negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.2 |
0.5 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 |
0.5 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.2 |
0.5 |
GO:0031627 |
telomeric loop formation(GO:0031627) |
| 0.2 |
0.5 |
GO:0042148 |
strand invasion(GO:0042148) |
| 0.2 |
0.5 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.2 |
0.5 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.2 |
0.7 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.2 |
0.3 |
GO:0046098 |
guanine metabolic process(GO:0046098) |
| 0.2 |
3.2 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 |
0.8 |
GO:0060295 |
regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 |
0.3 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 |
0.6 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 |
0.5 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 |
0.6 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.2 |
2.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
1.2 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 |
1.3 |
GO:0000972 |
transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 |
0.4 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.1 |
0.7 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 |
0.5 |
GO:0003347 |
epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.1 |
0.8 |
GO:0046502 |
uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 |
0.3 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) |
| 0.1 |
1.0 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.1 |
1.2 |
GO:0006183 |
GTP biosynthetic process(GO:0006183) |
| 0.1 |
0.4 |
GO:0021557 |
oculomotor nerve development(GO:0021557) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 |
0.9 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.1 |
0.7 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.1 |
0.4 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 |
0.4 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 |
0.4 |
GO:0034476 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 |
0.6 |
GO:1902856 |
negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 |
0.3 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.1 |
0.1 |
GO:0036503 |
ERAD pathway(GO:0036503) |
| 0.1 |
0.5 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 |
0.3 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 |
0.4 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 |
0.3 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
0.4 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 |
0.3 |
GO:0002765 |
immune response-inhibiting signal transduction(GO:0002765) |
| 0.1 |
0.4 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
1.0 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
1.4 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.1 |
0.5 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.1 |
0.6 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.1 |
0.5 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.1 |
0.7 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 |
0.6 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.1 |
0.5 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 |
0.5 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.1 |
0.5 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.1 |
0.5 |
GO:2000767 |
positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 |
0.5 |
GO:0045165 |
cell fate commitment(GO:0045165) |
| 0.1 |
3.4 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.1 |
0.3 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 |
0.3 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.1 |
0.6 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.3 |
GO:1900275 |
B cell proliferation involved in immune response(GO:0002322) negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 |
1.3 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
0.7 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 |
0.3 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.1 |
0.6 |
GO:0009162 |
nucleoside monophosphate catabolic process(GO:0009125) deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 |
0.3 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 |
0.7 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.1 |
0.4 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.1 |
0.3 |
GO:0032511 |
late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 |
0.3 |
GO:0036518 |
chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 |
0.5 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.1 |
0.2 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 |
1.6 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 |
1.1 |
GO:0090308 |
regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 |
0.2 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 |
0.1 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.1 |
0.5 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 |
0.7 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.1 |
0.4 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 |
0.5 |
GO:0007320 |
insemination(GO:0007320) |
| 0.1 |
0.5 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 |
0.1 |
GO:0042256 |
mature ribosome assembly(GO:0042256) |
| 0.1 |
0.3 |
GO:2001268 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 |
0.3 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.1 |
0.2 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.1 |
0.3 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.1 |
0.3 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 |
0.7 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 |
0.1 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.1 |
0.8 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
0.5 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
0.3 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 |
0.8 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.1 |
0.5 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
0.5 |
GO:0048318 |
axial mesoderm development(GO:0048318) |
| 0.1 |
0.5 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.1 |
0.4 |
GO:0070092 |
glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.1 |
0.2 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.1 |
0.2 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.1 |
0.2 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.1 |
0.2 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 |
0.5 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.1 |
0.4 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.1 |
0.5 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.4 |
GO:0034587 |
tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) piRNA metabolic process(GO:0034587) |
| 0.1 |
0.2 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.4 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 |
0.5 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 |
0.5 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 |
0.3 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.2 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
0.3 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 |
0.4 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 |
0.5 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
0.2 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.1 |
0.4 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 |
0.6 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 |
0.5 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
0.4 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.7 |
GO:0060252 |
positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.5 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 |
0.3 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
1.2 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.0 |
2.5 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.2 |
GO:2000195 |
negative regulation of female gonad development(GO:2000195) |
| 0.0 |
0.2 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 |
0.9 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.2 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
1.2 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.4 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.3 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.0 |
0.6 |
GO:0046685 |
response to arsenic-containing substance(GO:0046685) |
| 0.0 |
0.2 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 |
0.2 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 |
0.2 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.3 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.3 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.0 |
0.3 |
GO:0060315 |
negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 |
0.5 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.4 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
0.3 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.0 |
0.9 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.2 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.3 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 |
0.4 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.6 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 |
0.2 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.6 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.0 |
0.2 |
GO:0051096 |
regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.2 |
GO:0021798 |
forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 |
0.6 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.2 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 |
0.3 |
GO:0060371 |
regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 |
0.1 |
GO:0043129 |
surfactant homeostasis(GO:0043129) |
| 0.0 |
0.2 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.1 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.0 |
0.1 |
GO:0060523 |
prostate epithelial cord elongation(GO:0060523) |
| 0.0 |
0.2 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
0.4 |
GO:0042640 |
anagen(GO:0042640) |
| 0.0 |
1.9 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.4 |
GO:0046349 |
amino sugar biosynthetic process(GO:0046349) |
| 0.0 |
0.1 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 |
0.4 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.0 |
0.5 |
GO:0070498 |
interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 |
0.2 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.3 |
GO:1990173 |
regulation of establishment of protein localization to telomere(GO:0070203) protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.1 |
GO:0035989 |
tendon development(GO:0035989) |
| 0.0 |
0.1 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.0 |
0.4 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.9 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.8 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.5 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 |
0.5 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.9 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.8 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.3 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.3 |
GO:1902166 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 |
0.4 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
0.8 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.0 |
0.1 |
GO:0042088 |
T-helper 1 type immune response(GO:0042088) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.3 |
GO:0031999 |
negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 |
1.2 |
GO:0021884 |
forebrain neuron development(GO:0021884) |
| 0.0 |
0.1 |
GO:0015838 |
amino-acid betaine transport(GO:0015838) |
| 0.0 |
0.3 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
1.1 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 |
0.1 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 |
0.5 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.3 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.0 |
0.9 |
GO:0006826 |
iron ion transport(GO:0006826) |
| 0.0 |
0.7 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.1 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 |
0.4 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.5 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
0.5 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.1 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.2 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 |
0.2 |
GO:0060346 |
bone trabecula formation(GO:0060346) |
| 0.0 |
0.6 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.2 |
GO:0042559 |
folic acid-containing compound biosynthetic process(GO:0009396) pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 |
0.1 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 |
0.1 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 |
0.2 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.4 |
GO:0035067 |
negative regulation of histone acetylation(GO:0035067) |
| 0.0 |
0.4 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.2 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
1.3 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.0 |
0.0 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.0 |
1.1 |
GO:0002066 |
columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 |
0.2 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 |
0.2 |
GO:0044320 |
leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) |
| 0.0 |
0.4 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 |
0.4 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.1 |
GO:0016139 |
glycoside catabolic process(GO:0016139) |
| 0.0 |
0.6 |
GO:0072698 |
protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 |
0.1 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 |
0.5 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.2 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.3 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.0 |
0.1 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 |
1.2 |
GO:0042274 |
ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 |
0.2 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 |
0.1 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 |
0.2 |
GO:0048041 |
cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 |
0.3 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 |
0.3 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.4 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.6 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.2 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.2 |
GO:0042921 |
glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 |
0.9 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.0 |
2.1 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.1 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 |
0.2 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.0 |
0.3 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 |
0.9 |
GO:0019915 |
lipid storage(GO:0019915) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.4 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.0 |
0.1 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 |
0.1 |
GO:0031114 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.1 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.2 |
GO:0051092 |
positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 |
0.1 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 |
0.5 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.3 |
GO:0042438 |
melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.1 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.1 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.8 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.2 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.2 |
GO:0007519 |
skeletal muscle tissue development(GO:0007519) |
| 0.0 |
0.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.1 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.2 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 |
0.1 |
GO:0000466 |
maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 |
0.4 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.3 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 |
0.2 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.9 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.2 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.2 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.7 |
GO:0051289 |
protein homotetramerization(GO:0051289) |
| 0.0 |
0.3 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
0.1 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 |
1.1 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.4 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.4 |
GO:0006406 |
mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 |
0.1 |
GO:0036438 |
positive regulation of heterotypic cell-cell adhesion(GO:0034116) maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.4 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:0007096 |
regulation of exit from mitosis(GO:0007096) |
| 0.0 |
0.2 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 |
0.4 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 |
0.2 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.1 |
GO:0045862 |
positive regulation of proteolysis(GO:0045862) |
| 0.0 |
0.2 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.4 |
GO:0006919 |
activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 |
1.6 |
GO:0030178 |
negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 |
0.8 |
GO:0030518 |
intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 |
0.3 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 |
0.2 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.4 |
GO:0021549 |
cerebellum development(GO:0021549) |