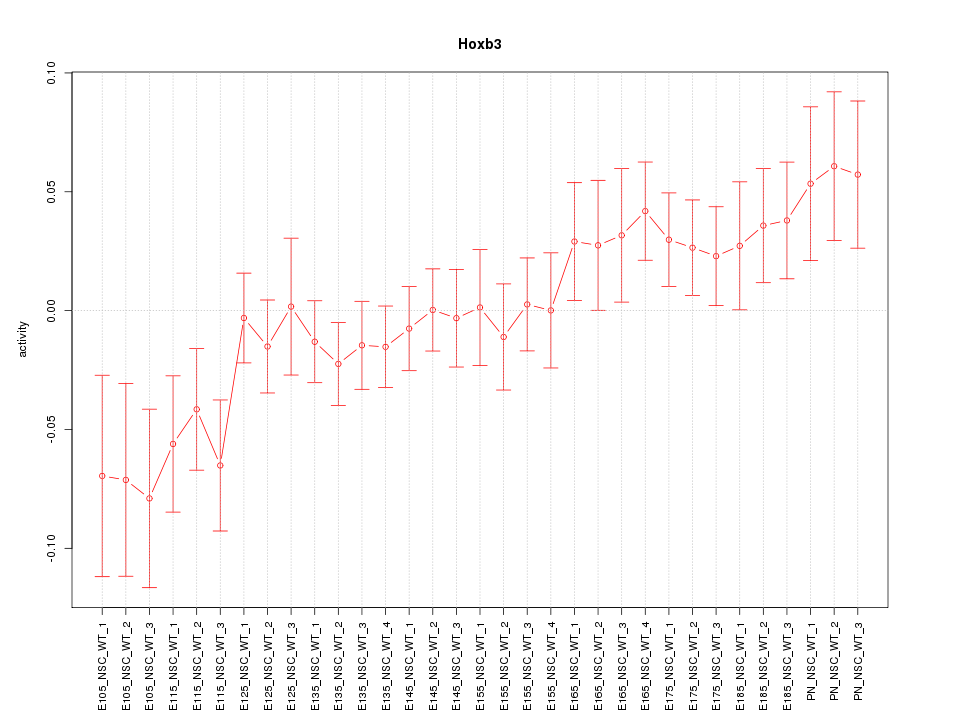
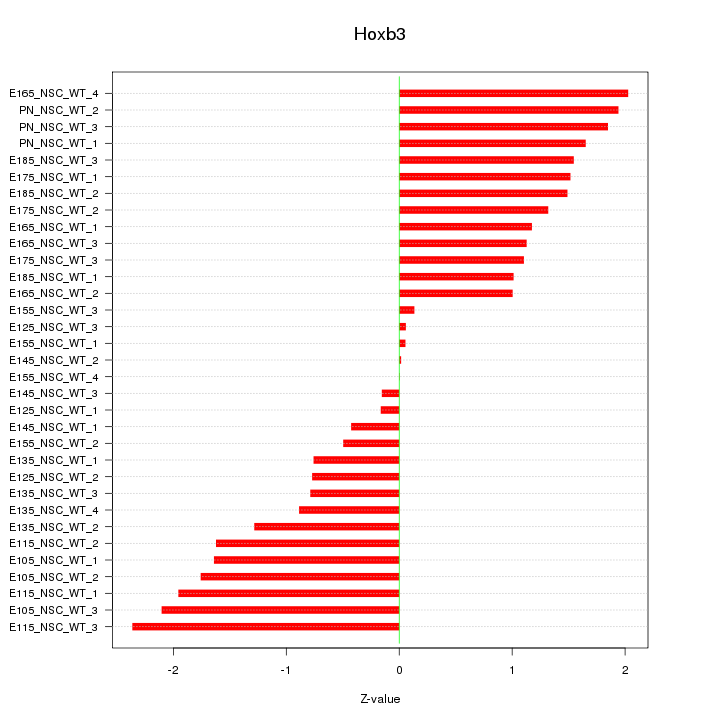
Motif ID: Hoxb3
Z-value: 1.298
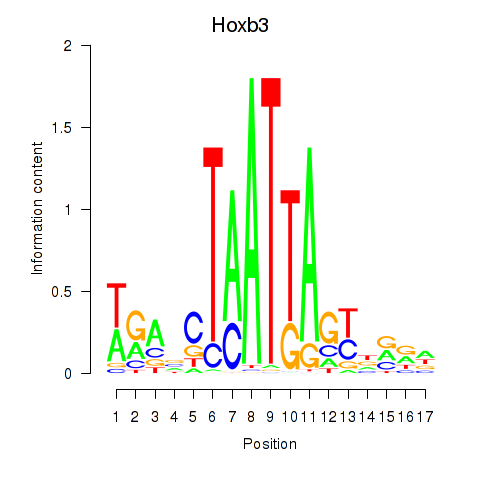
Transcription factors associated with Hoxb3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxb3 | ENSMUSG00000048763.5 | Hoxb3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 23.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.5 | 5.8 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.9 | 2.7 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.9 | 2.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.8 | 5.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.7 | 7.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.6 | 1.9 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.6 | 3.9 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.5 | 2.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 2.5 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.5 | 5.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.5 | 1.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.4 | 3.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 2.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 1.7 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.4 | 1.2 | GO:2000338 | positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.4 | 12.8 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.4 | 2.3 | GO:1903056 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 | 5.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 1.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.3 | 2.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 3.8 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 1.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 2.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 15.6 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.2 | 0.9 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 1.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.9 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 0.8 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.2 | 3.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 1.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.2 | 0.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 1.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 6.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.8 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 3.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 4.9 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 1.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.9 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.4 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.1 | 0.2 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 3.6 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 1.3 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.9 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 1.2 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 1.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 3.4 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 2.8 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.8 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.1 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) negative regulation of sequestering of calcium ion(GO:0051283) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 23.3 | GO:0043205 | fibril(GO:0043205) |
| 0.4 | 6.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 5.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 2.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 4.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 3.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 18.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 7.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 3.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 3.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 5.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 4.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 3.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 3.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 23.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.2 | 5.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 1.0 | 11.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.9 | 7.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.5 | 2.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 5.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 2.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 3.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 5.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.4 | 1.5 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.3 | 1.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.2 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.3 | 2.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 1.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 1.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 2.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.3 | 4.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 1.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 1.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 5.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 0.8 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 4.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 3.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 1.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 2.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 3.0 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.1 | 0.2 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.0 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 3.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 5.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0043024 | eukaryotic initiation factor 4E binding(GO:0008190) ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 2.7 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 1.2 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 1.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.8 | GO:0044325 | ion channel binding(GO:0044325) |