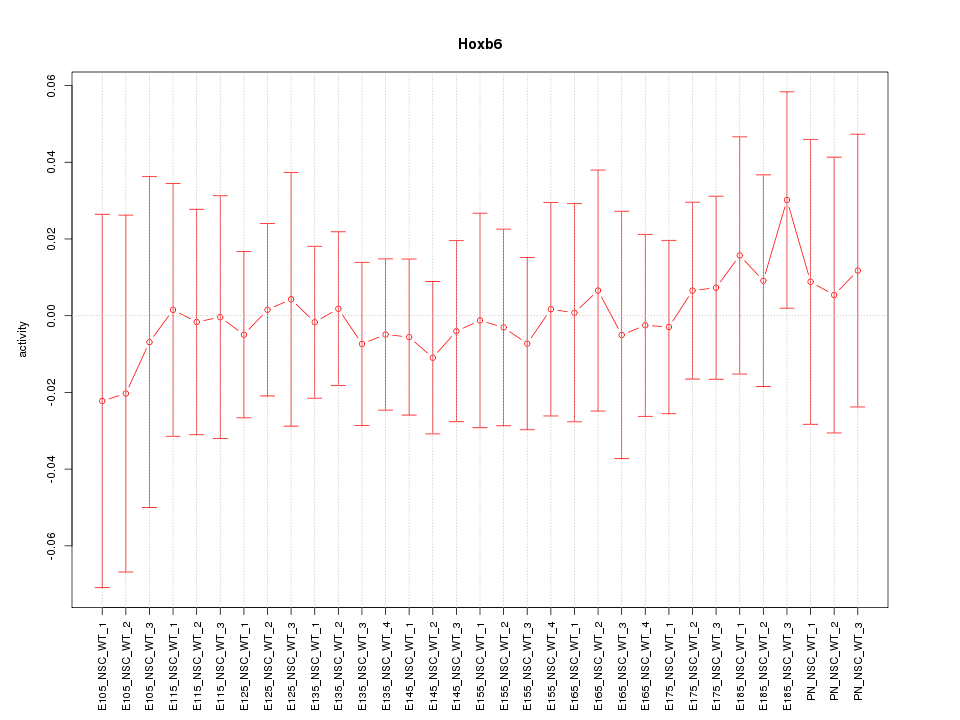
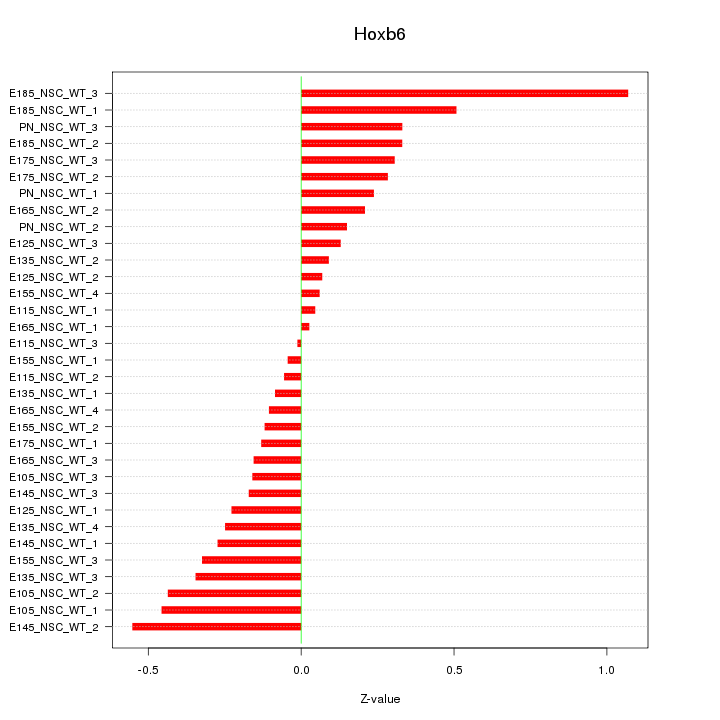
Motif ID: Hoxb6
Z-value: 0.312
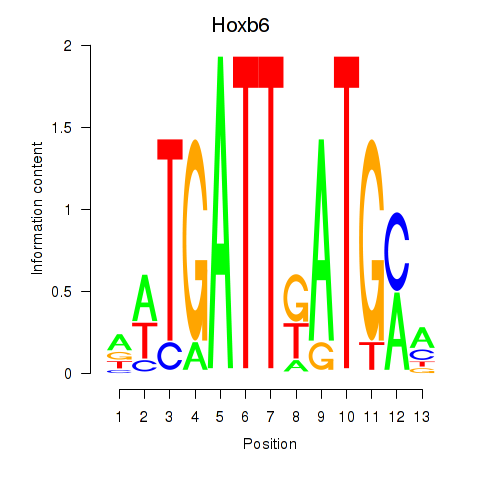
Transcription factors associated with Hoxb6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxb6 | ENSMUSG00000000690.4 | Hoxb6 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 0.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 0.5 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 1.3 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.5 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 1.6 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.9 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 1.3 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.7 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.3 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.1 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.4 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |