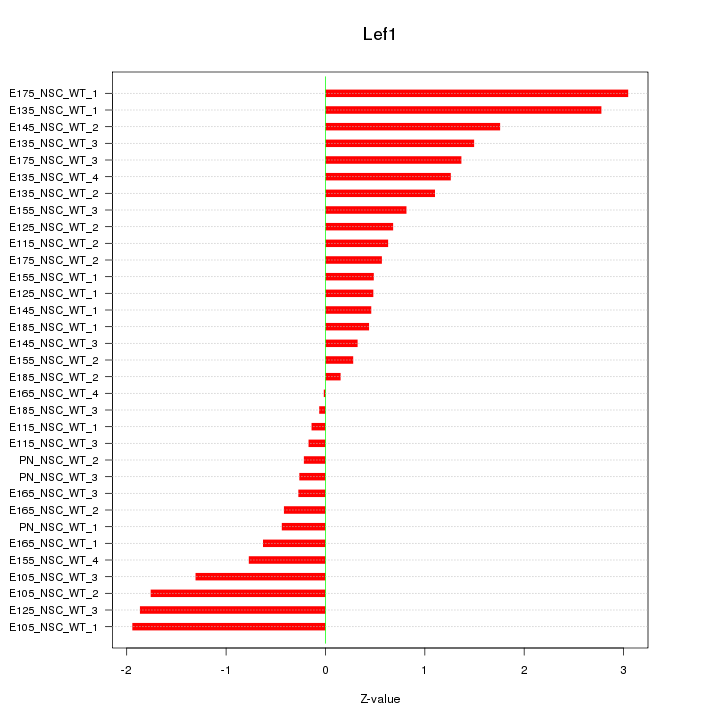
Motif ID: Lef1
Z-value: 1.149
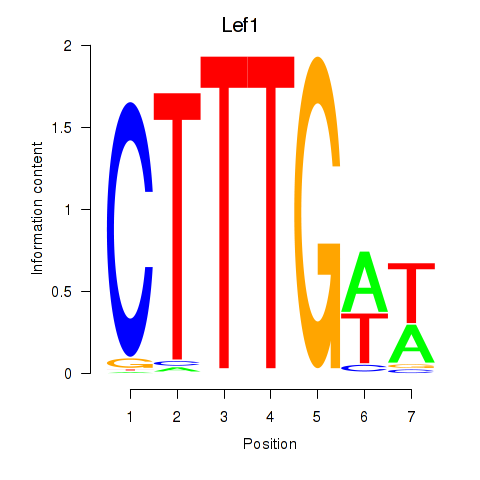
Transcription factors associated with Lef1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Lef1 | ENSMUSG00000027985.8 | Lef1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lef1 | mm10_v2_chr3_+_131112785_131112803 | -0.11 | 5.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.9 | 3.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.9 | 2.6 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.8 | 4.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.8 | 4.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.8 | 7.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.8 | 2.3 | GO:0048818 | positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.7 | 2.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.7 | 3.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 1.3 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.6 | 11.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.6 | 4.0 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.5 | 2.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.5 | 2.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.5 | 2.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.4 | 2.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.4 | 1.2 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.4 | 2.7 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.4 | 1.1 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 1.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.3 | 1.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.3 | 1.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 1.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.3 | 1.9 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.3 | 0.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 4.3 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 1.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 2.0 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.3 | 0.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 1.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.3 | 1.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 1.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 1.4 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 0.7 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 | 2.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 1.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 0.6 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 | 1.6 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 4.5 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.2 | 0.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.2 | 0.8 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.2 | 0.7 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 0.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 0.6 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.2 | 0.5 | GO:0002842 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 1.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.2 | 0.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.6 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.1 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 1.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.4 | GO:0050929 | corticospinal neuron axon guidance(GO:0021966) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.4 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.5 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 4.5 | GO:0071242 | cellular response to ammonium ion(GO:0071242) |
| 0.1 | 2.9 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 1.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 1.6 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 2.2 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 4.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.7 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 0.8 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.4 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 3.6 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.7 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 3.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 3.8 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 0.3 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 2.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 3.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.3 | GO:1905154 | negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 2.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 1.6 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.4 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 1.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.5 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 2.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.6 | GO:0035280 | miRNA mediated inhibition of translation(GO:0035278) miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.9 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.8 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 4.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.2 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 0.5 | GO:0086011 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) membrane repolarization during action potential(GO:0086011) regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 2.0 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.5 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.4 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.5 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 3.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.7 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.2 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.2 | GO:0060353 | cell adhesion molecule production(GO:0060352) regulation of cell adhesion molecule production(GO:0060353) renal water absorption(GO:0070295) |
| 0.0 | 0.4 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 1.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 2.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 2.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.9 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.6 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 3.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.2 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.7 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.8 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 1.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 2.3 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.5 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.2 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.4 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.1 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.5 | 2.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.4 | 3.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.4 | 7.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 3.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 3.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 4.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 4.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 3.8 | GO:0043205 | fibril(GO:0043205) |
| 0.2 | 1.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 0.6 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 3.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 3.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.8 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 3.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 3.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 2.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 1.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 20.5 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 5.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.0 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 1.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0001726 | ruffle(GO:0001726) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.8 | 3.8 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.6 | 2.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.5 | 2.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 1.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.4 | 1.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 1.2 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.4 | 1.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.4 | 9.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 7.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.4 | 1.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.3 | 1.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 4.0 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.3 | 5.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.2 | 2.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 4.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.2 | 2.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 0.8 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 3.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 1.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 3.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.6 | GO:0032795 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 2.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.8 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 2.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 2.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 6.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 3.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.8 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.4 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 6.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |