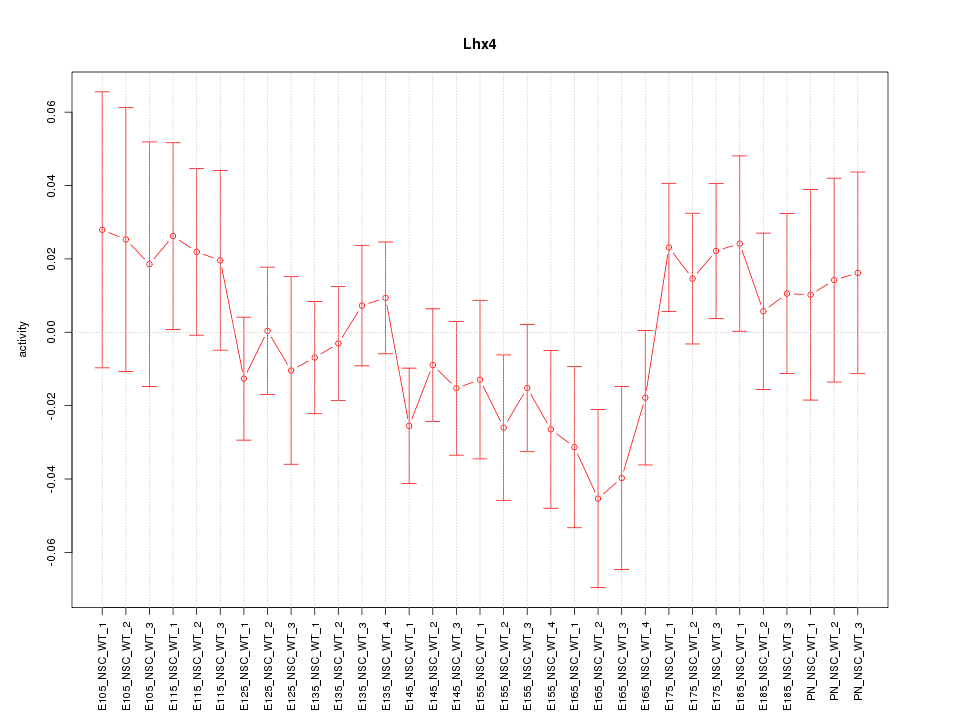
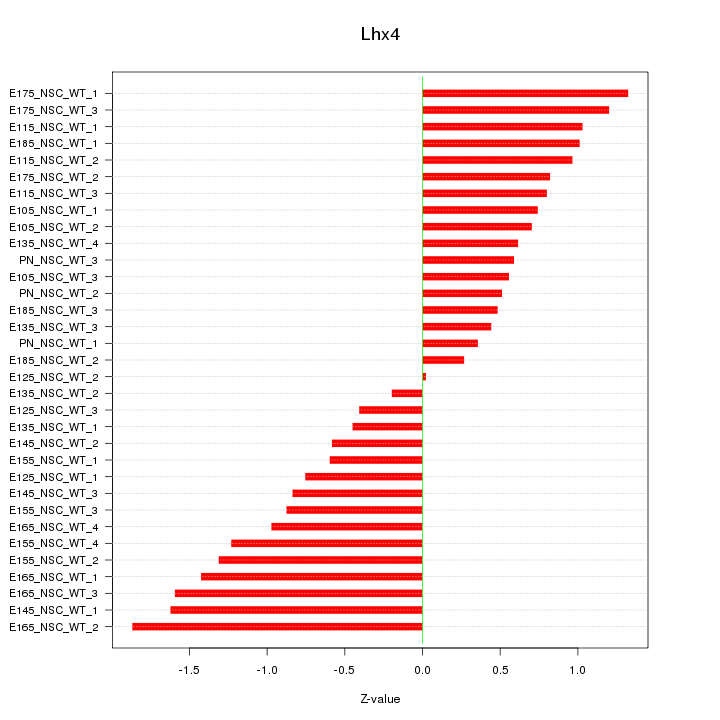
Motif ID: Lhx4
Z-value: 0.929
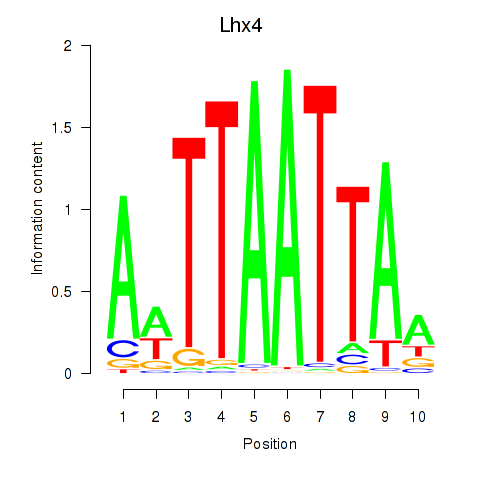
Transcription factors associated with Lhx4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Lhx4 | ENSMUSG00000026468.8 | Lhx4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.8 | 7.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.9 | 3.6 | GO:1900623 | positive regulation of keratinocyte proliferation(GO:0010838) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.9 | 2.7 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.8 | 3.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.7 | 2.2 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.7 | 2.1 | GO:0060423 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) negative regulation of mitotic cell cycle, embryonic(GO:0045976) foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.7 | 2.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.7 | 2.0 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.6 | 4.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.6 | 1.8 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.6 | 2.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.6 | 1.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.5 | 1.5 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.5 | 6.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.5 | 7.7 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.4 | 7.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 2.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.4 | 1.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.4 | 1.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 3.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.3 | 1.7 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 0.9 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.3 | 0.9 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 | 0.9 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 0.8 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.3 | 6.8 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.2 | 1.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 | 1.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 0.7 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 0.8 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 1.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 4.8 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.2 | 0.4 | GO:0046878 | regulation of saliva secretion(GO:0046877) positive regulation of saliva secretion(GO:0046878) |
| 0.2 | 0.7 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.2 | 1.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 1.7 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.2 | 4.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.6 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 2.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.4 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 2.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 2.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 4.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.7 | GO:0071803 | keratinocyte development(GO:0003334) positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 2.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 1.6 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 1.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.7 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 1.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.1 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.1 | 2.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.0 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 2.0 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 0.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 3.1 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 1.0 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 4.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 1.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 1.0 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.8 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.6 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 1.7 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.1 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 8.1 | GO:0016055 | Wnt signaling pathway(GO:0016055) cell-cell signaling by wnt(GO:0198738) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 1.4 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.7 | 2.2 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.3 | 2.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 1.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 1.0 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 1.7 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.2 | 3.3 | GO:0043205 | fibril(GO:0043205) |
| 0.2 | 0.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 0.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 6.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 2.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.7 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.0 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 14.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 2.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 6.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 2.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.9 | GO:0072372 | primary cilium(GO:0072372) |
| 0.0 | 1.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.5 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.2 | 3.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.9 | 2.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.7 | 4.9 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.7 | 3.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.5 | 2.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.5 | 7.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 1.4 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 2.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) neuregulin binding(GO:0038132) |
| 0.4 | 2.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 1.7 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.4 | 1.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.3 | 1.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 0.9 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.3 | 11.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 2.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.3 | 2.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 2.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 1.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.3 | 2.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.7 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.2 | 1.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.2 | 0.7 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.2 | 1.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.8 | GO:0051378 | primary amine oxidase activity(GO:0008131) serotonin binding(GO:0051378) |
| 0.2 | 6.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 2.5 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 5.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 4.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.6 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 7.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.8 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.0 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 1.0 | GO:0050811 | GABA-A receptor activity(GO:0004890) GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 1.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.5 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 2.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.8 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.7 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.0 | 1.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 6.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.0 | 1.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.8 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 3.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 3.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.1 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |