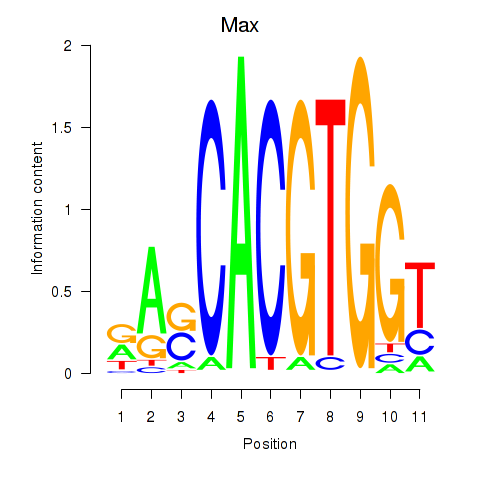
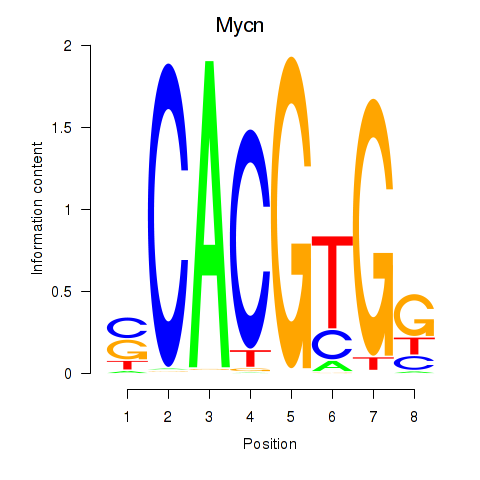
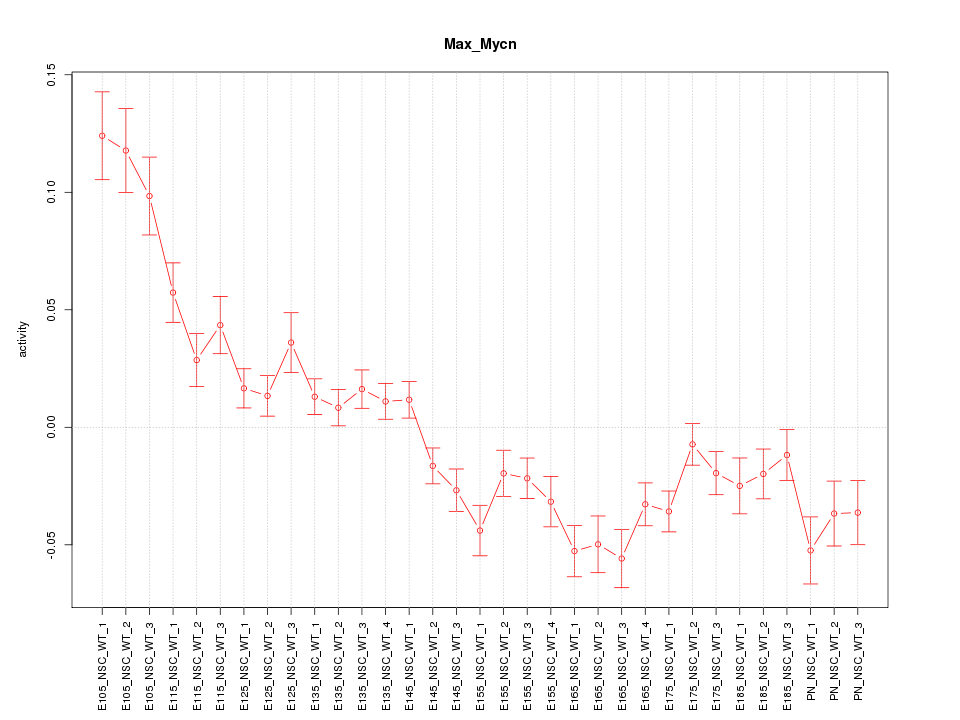
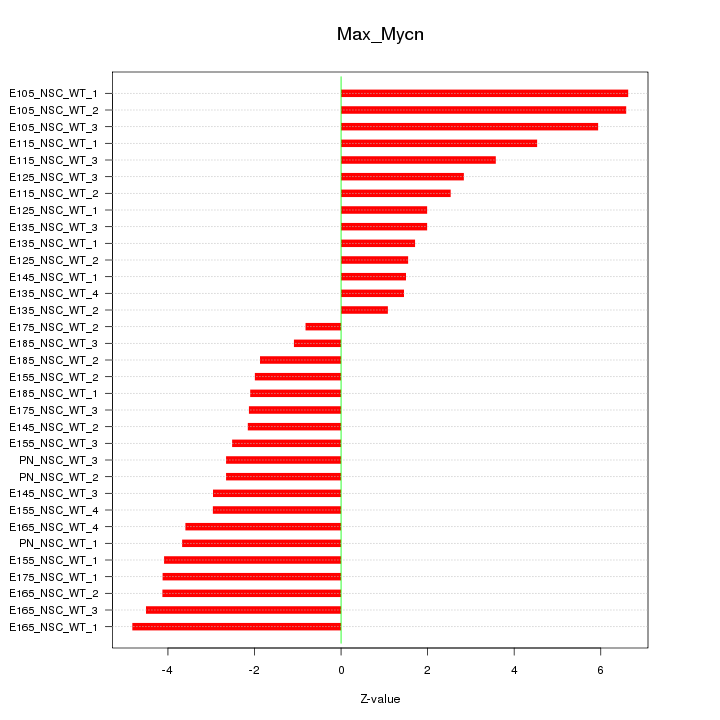
| 9.5 |
37.8 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 7.7 |
46.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 6.8 |
13.6 |
GO:0048389 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) ureter urothelium development(GO:0072190) |
| 6.5 |
32.3 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 6.2 |
30.9 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 6.2 |
18.5 |
GO:0086017 |
Purkinje myocyte action potential(GO:0086017) membrane depolarization during SA node cell action potential(GO:0086046) |
| 5.8 |
17.5 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 5.7 |
17.0 |
GO:0072180 |
mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 5.5 |
5.5 |
GO:0009153 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 5.4 |
16.2 |
GO:0006059 |
hexitol metabolic process(GO:0006059) |
| 5.0 |
15.1 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 5.0 |
15.0 |
GO:0009177 |
deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 4.6 |
22.9 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 4.5 |
18.2 |
GO:0070858 |
negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 4.4 |
21.8 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 4.4 |
13.1 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 4.3 |
12.9 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 4.1 |
8.3 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 4.0 |
12.0 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 4.0 |
4.0 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 3.9 |
7.9 |
GO:1905065 |
positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 3.7 |
3.7 |
GO:0072061 |
inner medullary collecting duct development(GO:0072061) |
| 3.7 |
18.6 |
GO:0019249 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 3.6 |
10.9 |
GO:0000965 |
mitochondrial RNA 3'-end processing(GO:0000965) |
| 3.6 |
10.8 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 3.6 |
10.8 |
GO:1904706 |
response to cisplatin(GO:0072718) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 3.5 |
3.5 |
GO:1901536 |
regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 3.4 |
17.1 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 3.4 |
30.4 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 3.3 |
6.6 |
GO:1900368 |
regulation of RNA interference(GO:1900368) |
| 3.3 |
39.7 |
GO:0009209 |
pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) |
| 3.2 |
19.1 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 3.2 |
12.7 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 3.1 |
15.6 |
GO:0040031 |
snRNA modification(GO:0040031) |
| 3.1 |
9.3 |
GO:0042908 |
xenobiotic transport(GO:0042908) |
| 3.0 |
3.0 |
GO:0060290 |
transdifferentiation(GO:0060290) |
| 3.0 |
9.0 |
GO:0009130 |
pyrimidine nucleoside monophosphate biosynthetic process(GO:0009130) |
| 3.0 |
11.9 |
GO:0090238 |
positive regulation of arachidonic acid secretion(GO:0090238) |
| 2.9 |
8.8 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 2.8 |
8.5 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 2.8 |
19.7 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 2.8 |
11.1 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 2.8 |
16.6 |
GO:0018195 |
peptidyl-arginine modification(GO:0018195) |
| 2.8 |
8.3 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 2.7 |
8.2 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 2.6 |
7.9 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 2.6 |
36.9 |
GO:0002192 |
IRES-dependent translational initiation(GO:0002192) |
| 2.6 |
7.8 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 2.6 |
7.8 |
GO:1903054 |
lysosomal membrane organization(GO:0097212) negative regulation of extracellular matrix organization(GO:1903054) positive regulation of protein folding(GO:1903334) |
| 2.6 |
5.2 |
GO:0010643 |
cell communication by chemical coupling(GO:0010643) |
| 2.6 |
10.3 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 2.6 |
10.3 |
GO:0061626 |
pharyngeal arch artery morphogenesis(GO:0061626) |
| 2.5 |
10.2 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 2.5 |
10.0 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 2.5 |
9.9 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 2.4 |
2.4 |
GO:0046084 |
adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 2.4 |
7.3 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 2.4 |
12.1 |
GO:0071038 |
nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 2.4 |
17.0 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 2.4 |
7.2 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 2.4 |
23.7 |
GO:0000466 |
maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 2.3 |
7.0 |
GO:0060437 |
lung growth(GO:0060437) trachea cartilage morphogenesis(GO:0060535) |
| 2.3 |
11.5 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 2.3 |
9.1 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 2.2 |
15.4 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 2.2 |
8.7 |
GO:0072369 |
regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 2.2 |
10.8 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 2.2 |
6.5 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 2.1 |
8.5 |
GO:1902570 |
protein localization to nucleolus(GO:1902570) |
| 2.1 |
6.3 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 2.1 |
6.3 |
GO:0006592 |
ornithine biosynthetic process(GO:0006592) |
| 2.1 |
4.1 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 2.0 |
14.1 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 2.0 |
2.0 |
GO:0016078 |
tRNA catabolic process(GO:0016078) |
| 2.0 |
9.9 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 2.0 |
9.8 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 1.9 |
5.8 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 1.9 |
1.9 |
GO:0035413 |
positive regulation of catenin import into nucleus(GO:0035413) |
| 1.9 |
7.8 |
GO:0070827 |
chromatin maintenance(GO:0070827) heterochromatin maintenance(GO:0070829) |
| 1.9 |
3.9 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 1.9 |
5.8 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.9 |
26.7 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 1.8 |
7.4 |
GO:0032439 |
endosome localization(GO:0032439) |
| 1.8 |
11.0 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 1.8 |
1.8 |
GO:0046131 |
pyrimidine ribonucleoside metabolic process(GO:0046131) |
| 1.8 |
7.3 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 1.8 |
5.4 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 1.8 |
5.4 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 1.8 |
7.1 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 1.8 |
36.8 |
GO:0006220 |
pyrimidine nucleotide metabolic process(GO:0006220) |
| 1.7 |
12.2 |
GO:0006855 |
drug transmembrane transport(GO:0006855) |
| 1.7 |
10.3 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 1.7 |
10.3 |
GO:0036089 |
cleavage furrow formation(GO:0036089) |
| 1.7 |
13.4 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 1.7 |
25.1 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 1.7 |
8.3 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 1.6 |
14.6 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 1.6 |
24.1 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 1.6 |
4.8 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 1.6 |
4.8 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 1.6 |
4.7 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.6 |
6.3 |
GO:0060032 |
notochord regression(GO:0060032) |
| 1.6 |
4.7 |
GO:0007354 |
zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 1.6 |
4.7 |
GO:0010899 |
regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 1.6 |
4.7 |
GO:0032053 |
ciliary basal body organization(GO:0032053) |
| 1.5 |
6.2 |
GO:0060266 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 1.5 |
4.6 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.5 |
3.0 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 1.5 |
7.6 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 1.5 |
4.5 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 1.5 |
9.0 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 1.5 |
4.5 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 1.5 |
8.9 |
GO:0033313 |
meiotic cell cycle checkpoint(GO:0033313) |
| 1.5 |
3.0 |
GO:0045343 |
MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 1.5 |
11.8 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 1.5 |
5.9 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 1.5 |
2.9 |
GO:0072071 |
mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 1.5 |
2.9 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 1.4 |
5.7 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 1.4 |
2.8 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 1.4 |
51.3 |
GO:0006414 |
translational elongation(GO:0006414) |
| 1.4 |
6.9 |
GO:0015705 |
iodide transport(GO:0015705) |
| 1.4 |
8.3 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 1.4 |
8.1 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 1.3 |
2.7 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 1.3 |
4.0 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 1.3 |
14.5 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 1.3 |
2.6 |
GO:0046370 |
fructose biosynthetic process(GO:0046370) |
| 1.3 |
10.5 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 1.3 |
3.9 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 1.3 |
5.2 |
GO:1902263 |
apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 1.3 |
15.6 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 1.3 |
3.9 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 1.3 |
3.9 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.3 |
5.2 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 1.3 |
9.0 |
GO:0001839 |
neural plate morphogenesis(GO:0001839) |
| 1.3 |
3.9 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 1.3 |
6.4 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 1.3 |
5.1 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.3 |
6.3 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 1.3 |
3.8 |
GO:0000237 |
leptotene(GO:0000237) |
| 1.2 |
5.0 |
GO:1904872 |
regulation of telomerase RNA localization to Cajal body(GO:1904872) positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 1.2 |
6.2 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.2 |
5.0 |
GO:2000195 |
negative regulation of female gonad development(GO:2000195) |
| 1.2 |
3.7 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 1.2 |
9.8 |
GO:0045056 |
transcytosis(GO:0045056) |
| 1.2 |
11.0 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 1.2 |
18.2 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 1.2 |
6.1 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 1.2 |
3.6 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 1.2 |
13.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 1.2 |
6.0 |
GO:0060066 |
oviduct development(GO:0060066) |
| 1.2 |
4.7 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 1.2 |
3.6 |
GO:0000727 |
double-strand break repair via break-induced replication(GO:0000727) |
| 1.2 |
7.0 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 1.2 |
12.9 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 1.2 |
8.1 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 1.2 |
3.5 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 1.2 |
10.4 |
GO:0033572 |
transferrin transport(GO:0033572) |
| 1.1 |
2.3 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 1.1 |
4.6 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 1.1 |
14.9 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 1.1 |
12.4 |
GO:0007440 |
foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 1.1 |
15.8 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 1.1 |
4.5 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 1.1 |
22.4 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 1.1 |
3.4 |
GO:0030214 |
hyaluronan catabolic process(GO:0030214) |
| 1.1 |
3.3 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 1.1 |
2.2 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 1.1 |
1.1 |
GO:1902109 |
negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 1.1 |
2.2 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 1.1 |
1.1 |
GO:0034287 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 1.1 |
4.4 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 1.1 |
3.3 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 1.1 |
4.3 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 1.1 |
4.3 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 1.1 |
6.4 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 1.1 |
2.1 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 1.1 |
3.2 |
GO:1904816 |
positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 1.1 |
2.1 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 1.1 |
8.5 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 1.0 |
23.1 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 1.0 |
10.5 |
GO:0015884 |
folic acid transport(GO:0015884) |
| 1.0 |
3.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.0 |
4.2 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 1.0 |
5.2 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 1.0 |
12.4 |
GO:0010586 |
miRNA metabolic process(GO:0010586) |
| 1.0 |
1.0 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 1.0 |
9.2 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 1.0 |
1.0 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 1.0 |
7.1 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 1.0 |
6.0 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 1.0 |
1.0 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 1.0 |
3.9 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 1.0 |
11.8 |
GO:0032211 |
negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 1.0 |
4.9 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 1.0 |
24.5 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 1.0 |
23.4 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 1.0 |
4.9 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 1.0 |
19.4 |
GO:0046033 |
AMP metabolic process(GO:0046033) |
| 1.0 |
1.9 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 1.0 |
2.9 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 1.0 |
3.8 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 1.0 |
4.8 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.9 |
6.6 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.9 |
4.7 |
GO:0010917 |
negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.9 |
7.4 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.9 |
3.7 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.9 |
12.9 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.9 |
1.8 |
GO:0031047 |
gene silencing by RNA(GO:0031047) |
| 0.9 |
2.7 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.9 |
5.4 |
GO:0032782 |
bile acid secretion(GO:0032782) |
| 0.9 |
4.5 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.9 |
36.5 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.9 |
8.0 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.9 |
4.4 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.9 |
2.6 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.9 |
4.3 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.9 |
3.5 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.9 |
1.7 |
GO:0031394 |
positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.9 |
3.4 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.9 |
1.7 |
GO:0033484 |
nitric oxide homeostasis(GO:0033484) |
| 0.8 |
2.5 |
GO:0046166 |
glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.8 |
8.5 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.8 |
12.6 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.8 |
5.8 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.8 |
6.7 |
GO:0009086 |
methionine biosynthetic process(GO:0009086) |
| 0.8 |
16.6 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.8 |
4.0 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.8 |
1.6 |
GO:0071034 |
CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.8 |
4.0 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.8 |
1.6 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.8 |
16.4 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.8 |
13.9 |
GO:0048255 |
mRNA stabilization(GO:0048255) |
| 0.8 |
5.4 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.8 |
3.8 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.8 |
0.8 |
GO:0003419 |
growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.8 |
3.0 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) positive regulation of male germ cell proliferation(GO:2000256) |
| 0.8 |
6.0 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.7 |
0.7 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.7 |
5.2 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.7 |
7.4 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.7 |
3.0 |
GO:0000255 |
allantoin metabolic process(GO:0000255) |
| 0.7 |
3.7 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.7 |
1.5 |
GO:0006109 |
regulation of carbohydrate metabolic process(GO:0006109) |
| 0.7 |
0.7 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.7 |
2.9 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) |
| 0.7 |
2.9 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.7 |
5.1 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.7 |
2.9 |
GO:0009435 |
NAD biosynthetic process(GO:0009435) |
| 0.7 |
1.5 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.7 |
1.4 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.7 |
4.3 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.7 |
2.1 |
GO:0014744 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of muscle adaptation(GO:0014744) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.7 |
2.8 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.7 |
2.1 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.7 |
2.1 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.7 |
2.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.7 |
2.8 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.7 |
1.4 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.7 |
4.1 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.7 |
5.5 |
GO:0042538 |
hyperosmotic salinity response(GO:0042538) |
| 0.7 |
9.5 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.7 |
18.3 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.7 |
2.0 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.7 |
8.7 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.7 |
1.3 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) |
| 0.7 |
14.5 |
GO:0006978 |
DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.7 |
14.5 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.7 |
2.0 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.6 |
2.6 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.6 |
7.7 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.6 |
6.4 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.6 |
1.9 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.6 |
8.9 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.6 |
7.6 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.6 |
6.3 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.6 |
1.9 |
GO:1904395 |
positive regulation of receptor clustering(GO:1903911) regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.6 |
18.9 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.6 |
7.5 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.6 |
11.9 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.6 |
6.9 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.6 |
3.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.6 |
3.7 |
GO:0036066 |
protein O-linked fucosylation(GO:0036066) |
| 0.6 |
3.6 |
GO:0046502 |
uroporphyrinogen III metabolic process(GO:0046502) |
| 0.6 |
34.9 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.6 |
3.0 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.6 |
9.5 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.6 |
3.0 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.6 |
1.2 |
GO:0006283 |
transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.6 |
2.9 |
GO:0090502 |
RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.6 |
11.7 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.6 |
2.3 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.6 |
1.2 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.6 |
1.7 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.6 |
3.5 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.6 |
12.0 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.6 |
0.6 |
GO:2000642 |
negative regulation of vacuolar transport(GO:1903336) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.6 |
1.1 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.6 |
1.7 |
GO:0030300 |
regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.6 |
1.1 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.6 |
1.7 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.6 |
5.1 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.6 |
1.7 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.6 |
4.5 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.6 |
5.6 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.6 |
34.8 |
GO:0006405 |
RNA export from nucleus(GO:0006405) |
| 0.6 |
2.8 |
GO:0051770 |
positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.5 |
5.5 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.5 |
2.2 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.5 |
1.6 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.5 |
6.0 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.5 |
1.1 |
GO:0043144 |
snoRNA processing(GO:0043144) |
| 0.5 |
1.6 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.5 |
1.6 |
GO:0015960 |
diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.5 |
4.3 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.5 |
12.4 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.5 |
7.5 |
GO:0042274 |
ribosomal small subunit biogenesis(GO:0042274) |
| 0.5 |
3.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.5 |
0.5 |
GO:0001844 |
protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.5 |
3.6 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.5 |
3.1 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.5 |
2.6 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.5 |
0.5 |
GO:0090110 |
cargo loading into COPII-coated vesicle(GO:0090110) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.5 |
2.0 |
GO:0071692 |
protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.5 |
3.1 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.5 |
5.1 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.5 |
3.0 |
GO:1901970 |
positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.5 |
2.0 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.5 |
8.9 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.5 |
1.5 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.5 |
3.4 |
GO:0033683 |
nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.5 |
25.8 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.5 |
7.1 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.5 |
0.5 |
GO:0033088 |
negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.5 |
0.9 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.5 |
11.3 |
GO:0060441 |
epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.5 |
3.3 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.5 |
3.8 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.5 |
2.4 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.5 |
4.7 |
GO:0090177 |
establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.5 |
0.9 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.5 |
1.4 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.5 |
2.8 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.5 |
2.3 |
GO:0019673 |
GDP-mannose metabolic process(GO:0019673) |
| 0.5 |
6.9 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.5 |
1.4 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.5 |
2.3 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.5 |
2.3 |
GO:2000157 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.4 |
3.6 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.4 |
4.0 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.4 |
0.9 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.4 |
4.9 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.4 |
7.5 |
GO:0014002 |
astrocyte development(GO:0014002) |
| 0.4 |
4.4 |
GO:0001773 |
myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.4 |
1.3 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.4 |
7.5 |
GO:0070286 |
axonemal dynein complex assembly(GO:0070286) |
| 0.4 |
12.7 |
GO:0030518 |
intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.4 |
2.2 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.4 |
1.3 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.4 |
7.8 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.4 |
1.7 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.4 |
4.3 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.4 |
1.3 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.4 |
3.0 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.4 |
3.8 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.4 |
5.5 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 |
1.7 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.4 |
1.2 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.4 |
0.8 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.4 |
1.6 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.4 |
7.3 |
GO:0006400 |
tRNA modification(GO:0006400) |
| 0.4 |
1.2 |
GO:0050929 |
corticospinal neuron axon guidance(GO:0021966) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.4 |
2.8 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.4 |
0.4 |
GO:2000277 |
positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.4 |
3.6 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.4 |
4.8 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 |
0.8 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.4 |
3.6 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.4 |
4.3 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.4 |
11.4 |
GO:0045880 |
positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.4 |
5.1 |
GO:0006670 |
sphingosine metabolic process(GO:0006670) |
| 0.4 |
1.6 |
GO:0060696 |
regulation of phospholipid catabolic process(GO:0060696) |
| 0.4 |
6.2 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.4 |
2.7 |
GO:0006490 |
oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.4 |
7.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.4 |
1.9 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.4 |
3.8 |
GO:0035067 |
negative regulation of histone acetylation(GO:0035067) |
| 0.4 |
1.1 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.4 |
5.3 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.4 |
1.9 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.4 |
2.2 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 |
0.7 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.4 |
1.1 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.4 |
5.2 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.4 |
3.3 |
GO:0019511 |
peptidyl-proline hydroxylation(GO:0019511) |
| 0.4 |
11.7 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.4 |
1.1 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.4 |
1.1 |
GO:0051096 |
regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.4 |
1.8 |
GO:0044854 |
plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.4 |
2.5 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.4 |
2.8 |
GO:0034123 |
positive regulation of toll-like receptor signaling pathway(GO:0034123) |
| 0.4 |
12.0 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.4 |
14.8 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.4 |
6.3 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.4 |
1.1 |
GO:0045080 |
positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.3 |
1.4 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.3 |
10.4 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.3 |
8.6 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 |
6.9 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.3 |
2.1 |
GO:0008611 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.3 |
22.4 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.3 |
1.4 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 |
1.7 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 |
7.3 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.3 |
1.6 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.3 |
1.9 |
GO:0071479 |
cellular response to ionizing radiation(GO:0071479) |
| 0.3 |
1.3 |
GO:0034115 |
negative regulation of heterotypic cell-cell adhesion(GO:0034115) cell-cell adhesion involved in gastrulation(GO:0070586) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.3 |
4.5 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 |
35.0 |
GO:0006626 |
protein targeting to mitochondrion(GO:0006626) |
| 0.3 |
1.0 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.3 |
8.2 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.3 |
4.3 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.3 |
1.5 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.3 |
0.9 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 |
6.4 |
GO:0001709 |
cell fate determination(GO:0001709) |
| 0.3 |
1.2 |
GO:0045176 |
asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.3 |
10.0 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.3 |
1.8 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.3 |
1.8 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.3 |
0.9 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.3 |
0.6 |
GO:0070445 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.3 |
1.2 |
GO:0010717 |
regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.3 |
1.8 |
GO:0003062 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.3 |
1.8 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 |
1.5 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.3 |
3.5 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.3 |
8.1 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 |
1.2 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.3 |
2.6 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.3 |
4.8 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.3 |
3.7 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.3 |
1.1 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.3 |
0.6 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 |
1.4 |
GO:0016560 |
protein import into peroxisome matrix, docking(GO:0016560) |
| 0.3 |
2.8 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.3 |
4.7 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.3 |
1.4 |
GO:0048563 |
post-embryonic organ morphogenesis(GO:0048563) |
| 0.3 |
3.3 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.3 |
1.1 |
GO:0030330 |
DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.3 |
2.4 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.3 |
2.1 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.3 |
2.6 |
GO:0003338 |
metanephros morphogenesis(GO:0003338) |
| 0.3 |
7.9 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.3 |
1.0 |
GO:0043550 |
regulation of lipid kinase activity(GO:0043550) |
| 0.3 |
1.0 |
GO:1904714 |
regulation of chaperone-mediated autophagy(GO:1904714) negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.3 |
3.3 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.3 |
2.0 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.2 |
9.7 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.2 |
5.2 |
GO:0006101 |
citrate metabolic process(GO:0006101) |
| 0.2 |
1.0 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 |
1.4 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 |
4.8 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.2 |
2.6 |
GO:0070585 |
protein localization to mitochondrion(GO:0070585) |
| 0.2 |
2.6 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.2 |
0.9 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 |
0.2 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.2 |
2.8 |
GO:0002052 |
positive regulation of neuroblast proliferation(GO:0002052) |
| 0.2 |
1.6 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.2 |
1.9 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 |
2.3 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 |
2.5 |
GO:0008637 |
apoptotic mitochondrial changes(GO:0008637) |
| 0.2 |
5.5 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.2 |
1.1 |
GO:1901030 |
positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.2 |
0.9 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.2 |
1.3 |
GO:0015817 |
histidine transport(GO:0015817) |
| 0.2 |
1.1 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.2 |
1.9 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.2 |
4.5 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.2 |
1.3 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.2 |
0.8 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 |
3.4 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.2 |
2.5 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 |
0.8 |
GO:0065001 |
negative regulation of histone H3-K36 methylation(GO:0000415) specification of axis polarity(GO:0065001) |
| 0.2 |
1.0 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.2 |
2.9 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.2 |
1.4 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 |
1.0 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.2 |
2.3 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.2 |
2.0 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 |
2.4 |
GO:0097150 |
neuronal stem cell population maintenance(GO:0097150) |
| 0.2 |
3.2 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.2 |
1.8 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.2 |
1.4 |
GO:0001942 |
hair follicle development(GO:0001942) molting cycle process(GO:0022404) hair cycle process(GO:0022405) skin epidermis development(GO:0098773) |
| 0.2 |
0.6 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.2 |
6.7 |
GO:0032755 |
positive regulation of interleukin-6 production(GO:0032755) |
| 0.2 |
20.5 |
GO:0006457 |
protein folding(GO:0006457) |
| 0.2 |
3.4 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 |
2.3 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 |
0.2 |
GO:1904526 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.2 |
0.4 |
GO:0030656 |
regulation of vitamin metabolic process(GO:0030656) |
| 0.2 |
0.6 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 |
0.4 |
GO:0061050 |
regulation of cell growth involved in cardiac muscle cell development(GO:0061050) positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.2 |
1.3 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.2 |
0.4 |
GO:0032753 |
positive regulation of interleukin-4 production(GO:0032753) |
| 0.2 |
2.8 |
GO:0014741 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.2 |
1.2 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 |
0.9 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.2 |
0.7 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 |
1.2 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.2 |
0.9 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.2 |
2.0 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 |
1.3 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.2 |
2.0 |
GO:0042461 |
photoreceptor cell development(GO:0042461) |
| 0.2 |
1.8 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 |
4.2 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.2 |
6.1 |
GO:0006418 |
tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 |
1.3 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.2 |
2.8 |
GO:0006310 |
DNA recombination(GO:0006310) |
| 0.2 |
0.8 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 |
2.7 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.2 |
2.6 |
GO:0048644 |
muscle organ morphogenesis(GO:0048644) |
| 0.2 |
2.9 |
GO:0033006 |
regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.2 |
0.5 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 |
3.4 |
GO:0045453 |
bone resorption(GO:0045453) |
| 0.2 |
1.4 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.2 |
0.9 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.2 |
1.8 |
GO:0050850 |
positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.2 |
0.8 |
GO:0060665 |
mesenchymal-epithelial cell signaling(GO:0060638) regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
10.6 |
GO:0002062 |
chondrocyte differentiation(GO:0002062) |
| 0.1 |
1.2 |
GO:0055012 |
ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.1 |
1.9 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.1 |
8.9 |
GO:2000045 |
regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.1 |
0.4 |
GO:0046016 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.1 |
0.6 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
1.7 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 |
1.7 |
GO:0007492 |
endoderm development(GO:0007492) |
| 0.1 |
0.4 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.1 |
3.0 |
GO:1902017 |
regulation of cilium assembly(GO:1902017) |
| 0.1 |
1.3 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 |
1.9 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.1 |
2.0 |
GO:0017038 |
protein import(GO:0017038) |
| 0.1 |
4.8 |
GO:0090305 |
nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.1 |
0.8 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 |
0.5 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
4.9 |
GO:0008033 |
tRNA processing(GO:0008033) |
| 0.1 |
1.8 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.1 |
1.1 |
GO:0002448 |
mast cell activation involved in immune response(GO:0002279) mast cell mediated immunity(GO:0002448) mast cell degranulation(GO:0043303) |
| 0.1 |
0.9 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
1.2 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.1 |
3.2 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.1 |
2.9 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 |
0.5 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 |
1.0 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 |
1.1 |
GO:0071451 |
cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.1 |
0.8 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 |
3.2 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 |
1.5 |
GO:0032760 |
positive regulation of tumor necrosis factor production(GO:0032760) positive regulation of tumor necrosis factor superfamily cytokine production(GO:1903557) |
| 0.1 |
5.4 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.1 |
0.3 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 |
1.2 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.1 |
8.9 |
GO:0045137 |
development of primary sexual characteristics(GO:0045137) |
| 0.1 |
2.2 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 |
0.5 |
GO:0003229 |
ventricular cardiac muscle tissue development(GO:0003229) |
| 0.1 |
0.8 |
GO:1900025 |
negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.1 |
1.3 |
GO:0034219 |
carbohydrate transmembrane transport(GO:0034219) |
| 0.1 |
2.3 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 |
6.7 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 0.1 |
9.3 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.1 |
1.6 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.1 |
2.0 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 |
2.2 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.1 |
0.4 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.1 |
0.6 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 |
0.3 |
GO:0001828 |
inner cell mass cell differentiation(GO:0001826) inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 |
2.3 |
GO:0019915 |
lipid storage(GO:0019915) |
| 0.1 |
0.7 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.6 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.9 |
GO:0002507 |
tolerance induction(GO:0002507) |
| 0.1 |
1.6 |
GO:0061647 |
histone H3-K9 modification(GO:0061647) |
| 0.1 |
1.4 |
GO:0042832 |
defense response to protozoan(GO:0042832) |
| 0.1 |
2.4 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.1 |
6.5 |
GO:0008286 |
insulin receptor signaling pathway(GO:0008286) |
| 0.1 |
0.6 |
GO:0097194 |
execution phase of apoptosis(GO:0097194) |
| 0.1 |
1.6 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 |
0.5 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.1 |
6.7 |
GO:0021915 |
neural tube development(GO:0021915) |
| 0.1 |
2.3 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.1 |
0.8 |
GO:0010666 |
positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 |
3.9 |
GO:0051297 |
centrosome organization(GO:0051297) |
| 0.1 |
0.5 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.1 |
2.2 |
GO:0002088 |
lens development in camera-type eye(GO:0002088) |
| 0.1 |
1.1 |
GO:0042921 |
glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 |
0.6 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 |
0.7 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 |
2.0 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.1 |
0.4 |
GO:0045862 |
positive regulation of proteolysis(GO:0045862) |
| 0.1 |
0.8 |
GO:0015858 |
nucleoside transport(GO:0015858) |
| 0.1 |
3.6 |
GO:0007286 |
spermatid development(GO:0007286) |
| 0.1 |
0.6 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.1 |
1.2 |
GO:0031954 |
positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 |
0.2 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 |
1.9 |
GO:2000648 |
positive regulation of stem cell proliferation(GO:2000648) |
| 0.1 |
1.8 |
GO:0035307 |
positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 |
0.3 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
1.5 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 |
1.1 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
2.4 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.1 |
0.3 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 |
1.2 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 |
0.8 |
GO:0000281 |
mitotic cytokinesis(GO:0000281) |
| 0.1 |
0.6 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
1.0 |
GO:0003382 |
epithelial cell morphogenesis(GO:0003382) |
| 0.1 |
0.7 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 |
0.5 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.1 |
0.3 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 |
0.5 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 |
0.5 |
GO:0008630 |
intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 |
0.5 |
GO:1901992 |
positive regulation of mitotic cell cycle phase transition(GO:1901992) |
| 0.1 |
0.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
1.2 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.1 |
0.2 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 |
0.7 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.1 |
0.2 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
0.1 |
GO:0051533 |
positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 |
1.8 |
GO:0051453 |
regulation of intracellular pH(GO:0051453) |
| 0.1 |
1.2 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.2 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 |
0.7 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 |
0.1 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.0 |
1.0 |
GO:1902653 |
cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 |
0.4 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.0 |
0.2 |
GO:0018027 |
peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 |
0.6 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.4 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.4 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 |
0.1 |
GO:0046040 |
IMP metabolic process(GO:0046040) |
| 0.0 |
0.2 |
GO:0009227 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 |
0.4 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
1.0 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
0.1 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 |
0.1 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 |
0.4 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.1 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 |
0.2 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.1 |
GO:0060468 |
prevention of polyspermy(GO:0060468) |
| 0.0 |
0.2 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.4 |
GO:0043267 |
negative regulation of potassium ion transport(GO:0043267) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.7 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.4 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.1 |
GO:0072337 |
modified amino acid transport(GO:0072337) |
| 0.0 |
0.7 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 |
0.1 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
5.0 |
GO:0006412 |
translation(GO:0006412) |
| 0.0 |
0.1 |
GO:0032026 |
response to magnesium ion(GO:0032026) |
| 0.0 |
0.1 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 |
0.0 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 |
0.3 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.3 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0071340 |
skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 |
0.3 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.1 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.5 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
0.3 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.1 |
GO:0018279 |
peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |