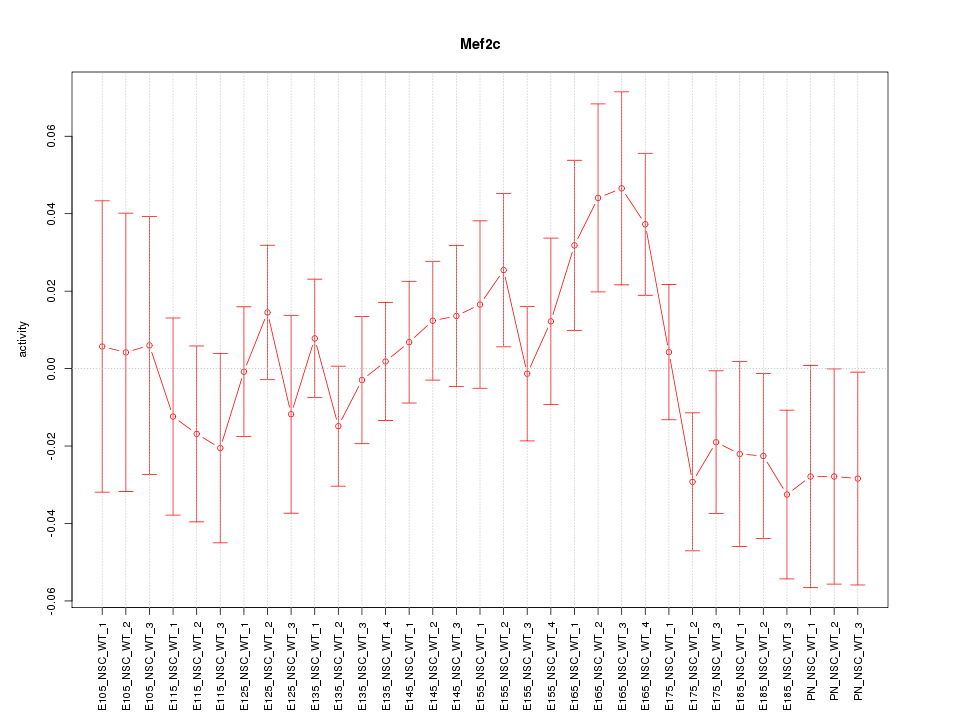
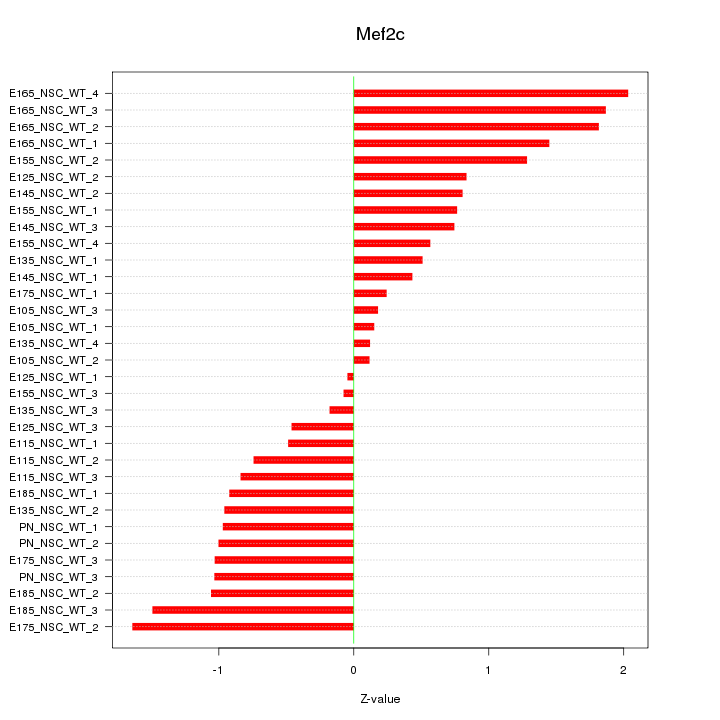
Motif ID: Mef2c
Z-value: 0.980
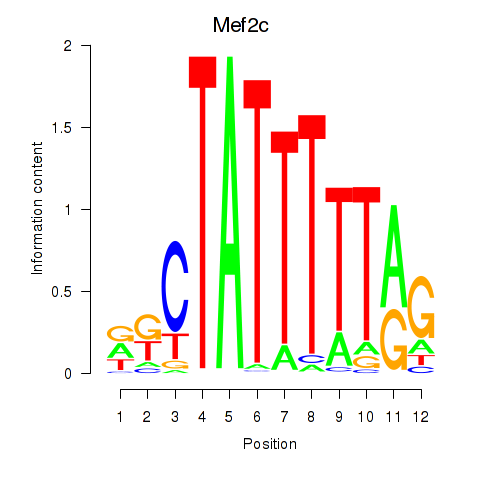
Transcription factors associated with Mef2c:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mef2c | ENSMUSG00000005583.10 | Mef2c |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2c | mm10_v2_chr13_+_83573577_83573607 | 0.63 | 9.4e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.5 | 4.5 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.5 | 16.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.8 | 4.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.7 | 2.9 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.7 | 2.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.5 | 2.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.5 | 2.4 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.5 | 1.4 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.4 | 5.8 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.4 | 1.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.4 | 3.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 2.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.3 | 1.0 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 1.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.3 | 2.7 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.3 | 3.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.3 | 2.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.3 | 4.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 1.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 3.9 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 1.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 0.9 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.2 | 4.6 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.2 | 1.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.2 | 0.4 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.2 | 0.8 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.2 | 0.8 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.2 | 0.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 5.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 3.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 2.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.9 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 3.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 2.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.4 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.5 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.1 | 0.7 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.1 | 0.4 | GO:0032511 | regulation of multivesicular body size(GO:0010796) late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 5.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 2.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.2 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.1 | 2.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 2.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.5 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 1.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 1.8 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 2.8 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 2.3 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 1.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 2.6 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 1.8 | GO:0051297 | centrosome organization(GO:0051297) |
| 0.0 | 1.2 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 1.1 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 2.8 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.9 | 2.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.8 | 16.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.7 | 4.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.5 | 1.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 5.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 0.9 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 1.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 0.5 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.2 | 1.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 2.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 4.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.8 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 5.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.2 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 7.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 3.9 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.7 | 4.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.7 | 4.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.6 | 2.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 16.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 1.5 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.4 | 4.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 2.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.4 | 1.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 4.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 0.9 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.3 | 9.7 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.3 | 5.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 2.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 2.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 3.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 4.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 1.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.7 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 1.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.8 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 3.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.8 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 2.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 2.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 8.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 2.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 3.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |