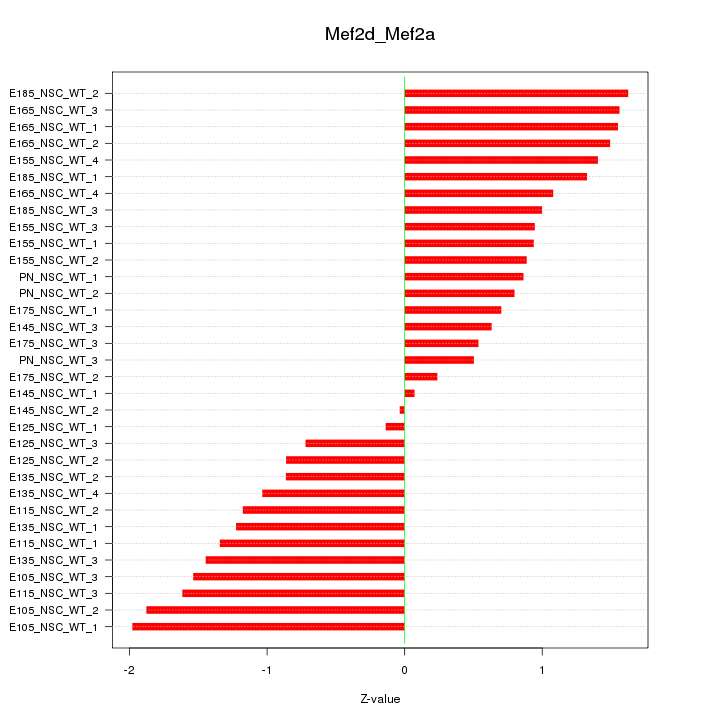
Motif ID: Mef2d_Mef2a
Z-value: 1.146
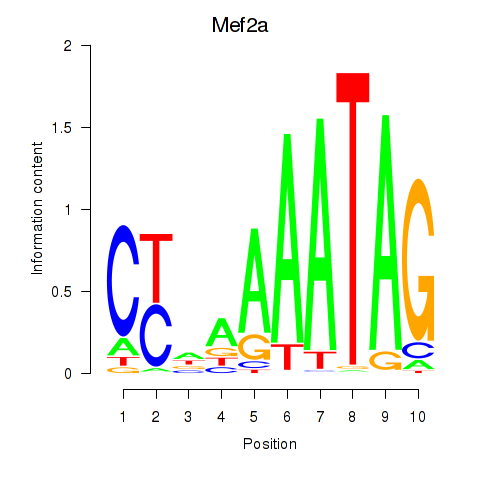
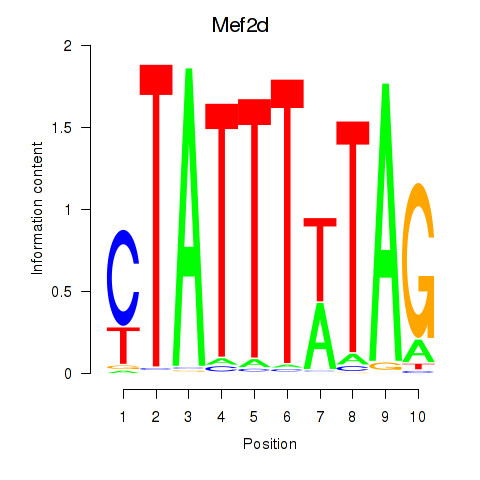
Transcription factors associated with Mef2d_Mef2a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mef2a | ENSMUSG00000030557.10 | Mef2a |
| Mef2d | ENSMUSG00000001419.11 | Mef2d |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2a | mm10_v2_chr7_-_67372846_67372858 | 0.95 | 1.0e-16 | Click! |
| Mef2d | mm10_v2_chr3_+_88142328_88142483 | 0.75 | 4.4e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 2.4 | 7.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 2.4 | 11.9 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 2.3 | 6.8 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 2.1 | 6.4 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 1.8 | 5.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 1.6 | 4.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.6 | 4.7 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 1.4 | 13.7 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 1.4 | 4.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 1.3 | 7.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 1.2 | 6.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.2 | 5.8 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.1 | 5.4 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 1.1 | 3.2 | GO:0048818 | positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 1.0 | 3.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.0 | 2.9 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 1.0 | 2.9 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 1.0 | 5.8 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.9 | 13.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.9 | 9.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.8 | 3.9 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.7 | 2.1 | GO:1903795 | regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.7 | 4.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.6 | 3.8 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.6 | 3.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.6 | 4.4 | GO:2000809 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.5 | 4.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.5 | 3.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 4.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.5 | 2.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 2.8 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.5 | 5.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.4 | 2.2 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.4 | 2.7 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.4 | 6.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.4 | 2.6 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.4 | 2.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 2.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.3 | 3.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 3.6 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 3.4 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.3 | 2.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.3 | 4.0 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.2 | 1.0 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 3.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 0.7 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.2 | 0.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 4.9 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.2 | 0.8 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.2 | 1.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.2 | 0.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.2 | 1.9 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) cellular response to magnesium ion(GO:0071286) regulation of membrane depolarization during action potential(GO:0098902) regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.5 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 3.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 5.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 14.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 1.9 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 1.7 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 1.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 4.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 1.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.8 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.1 | 3.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 7.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 7.7 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.8 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 3.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 3.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 1.8 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.7 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.9 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.5 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.1 | 2.1 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.1 | 4.1 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.1 | 0.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 1.8 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.6 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.9 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 2.4 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.6 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.6 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 2.2 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 4.9 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.3 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 4.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 3.9 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 1.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 2.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.7 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.3 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.2 | 8.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 1.1 | 4.3 | GO:0090537 | CERF complex(GO:0090537) |
| 1.0 | 2.9 | GO:0000802 | transverse filament(GO:0000802) |
| 0.9 | 6.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.9 | 4.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.8 | 15.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 2.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.5 | 4.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 2.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.4 | 7.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.4 | 2.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 5.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 1.0 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.3 | 6.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 6.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 5.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 3.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 8.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 3.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.4 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 2.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 22.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 4.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 5.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 3.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 5.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 0.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 3.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 6.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 5.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 3.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 6.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 4.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 4.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 4.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 4.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.7 | 11.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.5 | 6.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.4 | 4.3 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 1.2 | 8.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 1.1 | 3.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.0 | 7.1 | GO:0032795 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.9 | 13.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.8 | 4.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.8 | 6.4 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) steroid hormone binding(GO:1990239) |
| 0.8 | 4.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.8 | 3.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.6 | 5.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 2.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 5.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 1.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 4.0 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.4 | 7.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.3 | 1.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.3 | 1.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.3 | 6.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 5.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 5.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 2.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 2.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 1.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 2.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 2.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 1.9 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.2 | 10.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 3.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 5.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 0.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 4.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 2.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.7 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 3.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 2.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.8 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 4.0 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.1 | 4.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 5.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 4.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 1.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 6.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 2.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 3.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.5 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 9.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) cation:cation antiporter activity(GO:0015491) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 5.9 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.0 | 0.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 3.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |