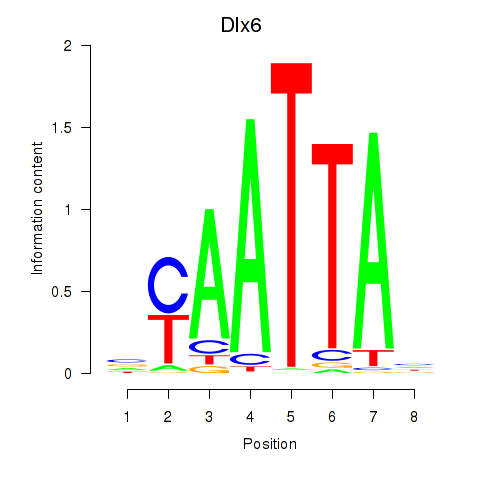
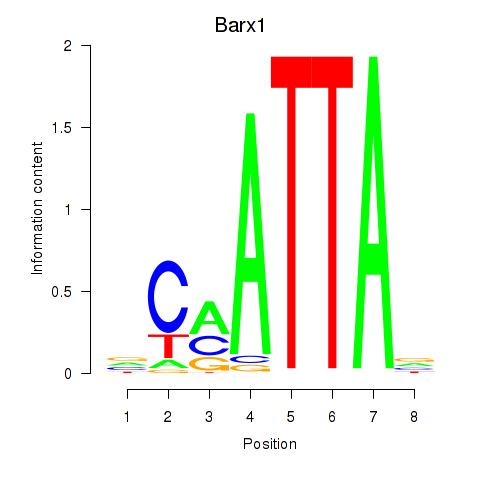
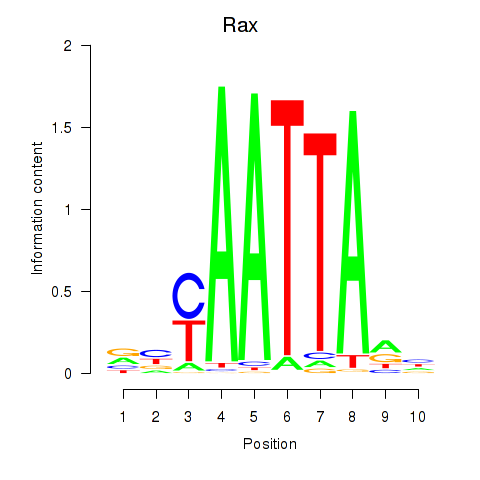
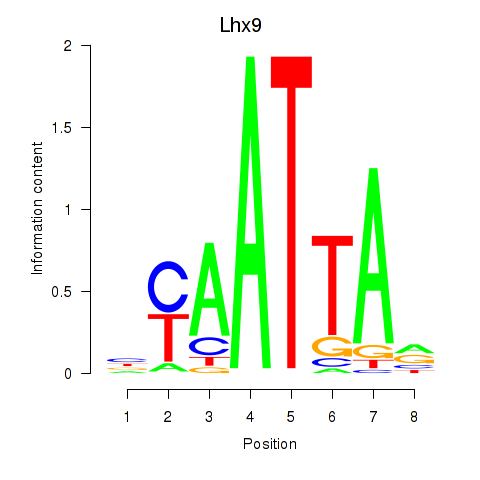
| 2.0 |
6.0 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.9 |
5.4 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.5 |
3.2 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.4 |
1.2 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.4 |
1.2 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 |
1.3 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.3 |
1.8 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.3 |
1.5 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 |
0.8 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.2 |
5.6 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.2 |
1.1 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.2 |
1.9 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 |
3.1 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.2 |
1.2 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.2 |
1.1 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 |
1.0 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
1.5 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.5 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.5 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
1.1 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.3 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 |
0.3 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.1 |
0.2 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.2 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 |
2.8 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.1 |
0.5 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.3 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
1.2 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
0.8 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.1 |
0.2 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 |
0.3 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) axonogenesis involved in innervation(GO:0060385) facioacoustic ganglion development(GO:1903375) |
| 0.1 |
0.2 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 |
0.8 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.1 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 |
0.8 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
1.6 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
0.1 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.0 |
0.6 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
3.6 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 |
0.2 |
GO:0097460 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.0 |
0.6 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.6 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
1.1 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.5 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.7 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.2 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.6 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 |
0.2 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.1 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
1.6 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.3 |
GO:0044253 |
positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 |
0.3 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.1 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.0 |
0.6 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
1.4 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
1.8 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
0.1 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
0.2 |
GO:2000249 |
negative regulation of cell-matrix adhesion(GO:0001953) regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 |
0.1 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |