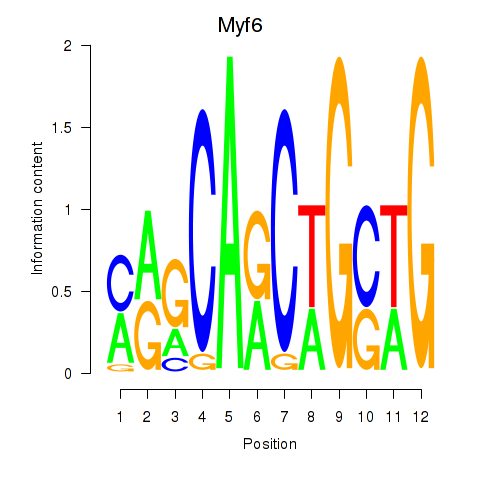
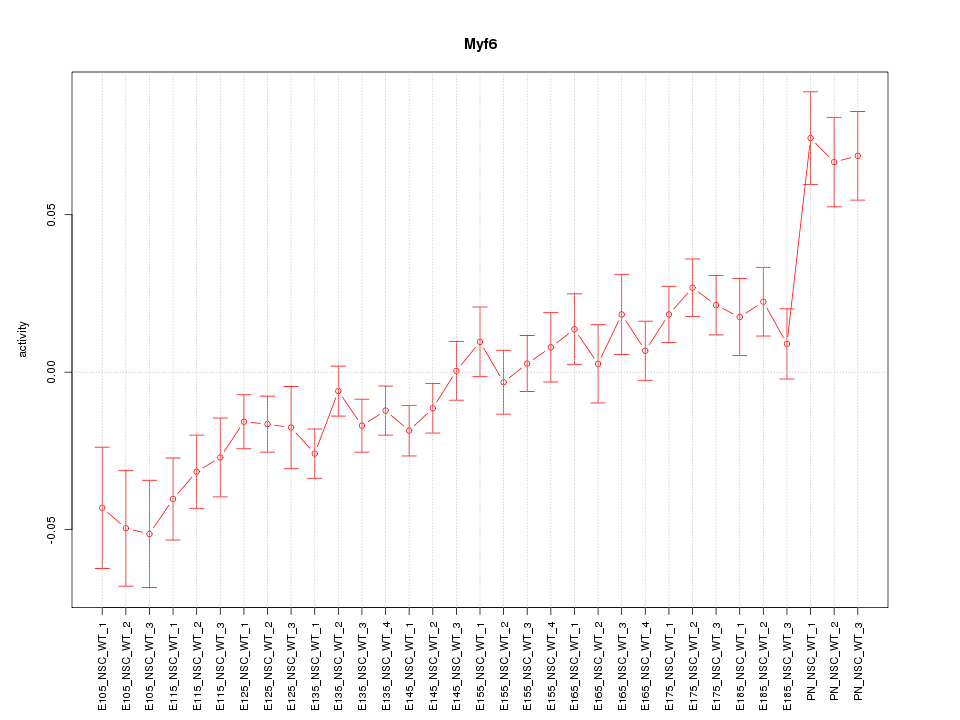
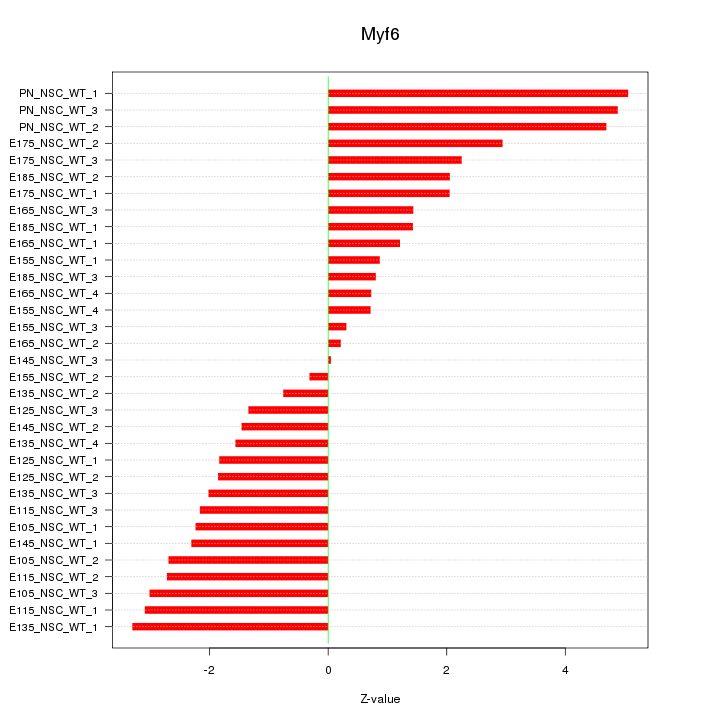
| 4.2 |
16.9 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 3.2 |
9.6 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 2.8 |
8.5 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 2.7 |
13.7 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 2.6 |
31.6 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 2.6 |
7.8 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 2.5 |
27.9 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 2.5 |
7.5 |
GO:0097212 |
lysosomal membrane organization(GO:0097212) |
| 2.5 |
12.4 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 2.4 |
4.9 |
GO:0035696 |
monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 2.4 |
7.1 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 2.4 |
14.1 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 2.1 |
6.4 |
GO:1900673 |
olefin metabolic process(GO:1900673) |
| 2.1 |
6.2 |
GO:0046959 |
habituation(GO:0046959) |
| 2.0 |
14.0 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.9 |
5.8 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 1.9 |
5.6 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 1.8 |
28.9 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 1.8 |
5.3 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 1.8 |
5.3 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 1.7 |
5.1 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.5 |
7.7 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 1.5 |
9.2 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 1.5 |
4.4 |
GO:0046544 |
development of secondary male sexual characteristics(GO:0046544) |
| 1.4 |
10.0 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 1.4 |
4.1 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.4 |
4.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 1.3 |
6.7 |
GO:0015871 |
choline transport(GO:0015871) |
| 1.3 |
4.0 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 1.3 |
5.2 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 1.3 |
3.9 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 1.2 |
4.7 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.2 |
11.6 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.2 |
4.6 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 1.1 |
3.4 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.1 |
4.5 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.1 |
3.3 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 1.1 |
17.3 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 1.0 |
4.2 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 1.0 |
10.8 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 1.0 |
5.9 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 1.0 |
5.8 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 1.0 |
2.9 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 1.0 |
2.9 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 1.0 |
2.9 |
GO:1902953 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.9 |
3.6 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.9 |
0.9 |
GO:0021593 |
rhombomere morphogenesis(GO:0021593) |
| 0.9 |
5.3 |
GO:1905206 |
positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.9 |
3.5 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.9 |
4.3 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.8 |
7.4 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.8 |
13.9 |
GO:0001964 |
startle response(GO:0001964) |
| 0.8 |
7.2 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.8 |
4.0 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.8 |
7.8 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.8 |
3.9 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.8 |
9.3 |
GO:0097084 |
vascular smooth muscle cell development(GO:0097084) |
| 0.8 |
3.1 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.8 |
7.5 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.7 |
2.9 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.7 |
10.5 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.7 |
2.1 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.7 |
2.1 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.7 |
6.1 |
GO:0060373 |
regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.6 |
2.6 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.6 |
1.9 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.6 |
4.4 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.6 |
2.5 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.6 |
2.2 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.6 |
1.1 |
GO:0051464 |
positive regulation of cortisol secretion(GO:0051464) |
| 0.5 |
4.1 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.5 |
3.1 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.5 |
0.5 |
GO:0086068 |
Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.5 |
2.0 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) lateral motor column neuron migration(GO:0097477) |
| 0.5 |
1.9 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.5 |
8.2 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.5 |
7.7 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.5 |
1.8 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.4 |
2.7 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.4 |
2.6 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.4 |
1.3 |
GO:0002586 |
positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.4 |
1.8 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.4 |
3.0 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.4 |
9.0 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.4 |
3.8 |
GO:0071435 |
potassium ion export(GO:0071435) |
| 0.4 |
1.3 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.4 |
1.3 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.4 |
3.0 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.4 |
10.3 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.4 |
5.4 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 |
2.0 |
GO:0006971 |
hypotonic response(GO:0006971) |
| 0.4 |
1.2 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.4 |
3.5 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 |
1.2 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.4 |
2.7 |
GO:0014053 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.4 |
1.5 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.4 |
0.4 |
GO:0072070 |
loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.4 |
1.5 |
GO:0050923 |
regulation of negative chemotaxis(GO:0050923) |
| 0.4 |
1.8 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 |
1.0 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.3 |
1.0 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.3 |
0.6 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.3 |
1.6 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.3 |
0.3 |
GO:0032916 |
transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.3 |
3.7 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.3 |
2.4 |
GO:0060536 |
cartilage morphogenesis(GO:0060536) |
| 0.3 |
3.2 |
GO:0045187 |
regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.3 |
9.1 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.3 |
2.6 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.3 |
7.8 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.3 |
5.1 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.3 |
2.1 |
GO:0010807 |
regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 |
0.8 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.3 |
3.6 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 |
4.9 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.3 |
1.0 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.3 |
1.3 |
GO:0033700 |
phospholipid efflux(GO:0033700) |
| 0.3 |
1.3 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.3 |
0.8 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.3 |
9.8 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 |
3.5 |
GO:1902254 |
negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.2 |
1.2 |
GO:0035188 |
blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 |
1.2 |
GO:0033690 |
positive regulation of osteoblast proliferation(GO:0033690) |
| 0.2 |
1.2 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.2 |
1.0 |
GO:0050913 |
sensory perception of bitter taste(GO:0050913) |
| 0.2 |
2.4 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.2 |
3.4 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.2 |
0.7 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 |
1.6 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 |
1.3 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 |
1.3 |
GO:0032055 |
negative regulation of translation in response to stress(GO:0032055) cellular response to cold(GO:0070417) |
| 0.2 |
1.1 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 |
1.1 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 |
1.1 |
GO:0002634 |
regulation of germinal center formation(GO:0002634) |
| 0.2 |
0.6 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.2 |
1.3 |
GO:0099625 |
regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.2 |
1.5 |
GO:0016139 |
glycoside catabolic process(GO:0016139) |
| 0.2 |
1.2 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 |
0.6 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) regulation of vascular wound healing(GO:0061043) |
| 0.2 |
1.0 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.2 |
2.6 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 |
2.5 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 |
1.1 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 |
3.4 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 |
0.4 |
GO:0035878 |
nail development(GO:0035878) |
| 0.2 |
2.4 |
GO:0046519 |
sphingoid metabolic process(GO:0046519) |
| 0.2 |
18.2 |
GO:0048709 |
oligodendrocyte differentiation(GO:0048709) |
| 0.2 |
1.6 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.2 |
5.2 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.2 |
2.2 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.2 |
2.6 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 |
7.7 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.2 |
0.3 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 |
0.7 |
GO:0010831 |
positive regulation of myotube differentiation(GO:0010831) |
| 0.2 |
4.2 |
GO:0009268 |
response to pH(GO:0009268) |
| 0.2 |
0.8 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.2 |
1.7 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 |
4.8 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 |
28.5 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.2 |
6.4 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.2 |
6.4 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.2 |
2.1 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 |
0.8 |
GO:1903779 |
regulation of cardiac conduction(GO:1903779) |
| 0.2 |
0.7 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.2 |
1.1 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 |
1.3 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.2 |
1.4 |
GO:0071578 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) zinc II ion transmembrane import(GO:0071578) |
| 0.1 |
0.7 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.1 |
9.3 |
GO:0030279 |
negative regulation of ossification(GO:0030279) |
| 0.1 |
3.5 |
GO:0051926 |
negative regulation of calcium ion transport(GO:0051926) |
| 0.1 |
0.4 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.1 |
3.2 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.1 |
2.0 |
GO:0044872 |
lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 |
0.9 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
6.6 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 |
13.0 |
GO:0030326 |
embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.1 |
2.0 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 |
1.1 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
0.2 |
GO:0042264 |
peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 |
0.4 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.1 |
0.3 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.1 |
1.0 |
GO:0046716 |
muscle cell cellular homeostasis(GO:0046716) |
| 0.1 |
1.9 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.1 |
1.2 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.1 |
1.0 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 |
5.3 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.1 |
2.1 |
GO:0001505 |
regulation of neurotransmitter levels(GO:0001505) |
| 0.1 |
0.7 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 |
1.7 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.1 |
0.4 |
GO:0048318 |
axial mesoderm development(GO:0048318) |
| 0.1 |
0.6 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
1.9 |
GO:0048546 |
digestive tract morphogenesis(GO:0048546) |
| 0.1 |
0.8 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 |
0.6 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.1 |
0.9 |
GO:1900273 |
positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.1 |
0.2 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 |
1.7 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.1 |
0.3 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 |
0.9 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.1 |
2.1 |
GO:0045921 |
positive regulation of exocytosis(GO:0045921) |
| 0.1 |
2.6 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 |
0.6 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 |
0.2 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.1 |
1.0 |
GO:0060292 |
long term synaptic depression(GO:0060292) |
| 0.1 |
0.3 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.1 |
0.4 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
1.0 |
GO:0001503 |
ossification(GO:0001503) |
| 0.1 |
1.2 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.6 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
1.9 |
GO:1902668 |
negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 |
1.1 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.1 |
GO:2000370 |
positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 |
2.2 |
GO:0007173 |
epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 |
1.0 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
2.3 |
GO:0035773 |
insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
| 0.0 |
0.8 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.0 |
0.5 |
GO:0051898 |
negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 |
4.0 |
GO:0098656 |
anion transmembrane transport(GO:0098656) |
| 0.0 |
2.5 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.0 |
2.3 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.0 |
0.5 |
GO:0003417 |
growth plate cartilage development(GO:0003417) |
| 0.0 |
0.8 |
GO:0086010 |
membrane depolarization during action potential(GO:0086010) |
| 0.0 |
0.7 |
GO:0019430 |
removal of superoxide radicals(GO:0019430) cellular oxidant detoxification(GO:0098869) |
| 0.0 |
1.8 |
GO:0048286 |
lung alveolus development(GO:0048286) |
| 0.0 |
1.0 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.5 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.8 |
GO:0006874 |
cellular calcium ion homeostasis(GO:0006874) |
| 0.0 |
2.3 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 |
0.2 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.1 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 |
0.1 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
1.2 |
GO:1902653 |
cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 |
0.2 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
3.5 |
GO:0006836 |
neurotransmitter transport(GO:0006836) |
| 0.0 |
0.2 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.4 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.0 |
0.2 |
GO:0001996 |
regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 |
2.9 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.2 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.8 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
0.7 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.0 |
1.4 |
GO:0043473 |
pigmentation(GO:0043473) |
| 0.0 |
2.9 |
GO:0006897 |
endocytosis(GO:0006897) |
| 0.0 |
0.5 |
GO:0048489 |
synaptic vesicle transport(GO:0048489) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 |
2.9 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
2.2 |
GO:0030010 |
establishment of cell polarity(GO:0030010) |
| 0.0 |
0.3 |
GO:0006744 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.1 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.0 |
0.1 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 |
0.3 |
GO:0042177 |
negative regulation of protein catabolic process(GO:0042177) |
| 0.0 |
0.3 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.3 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |