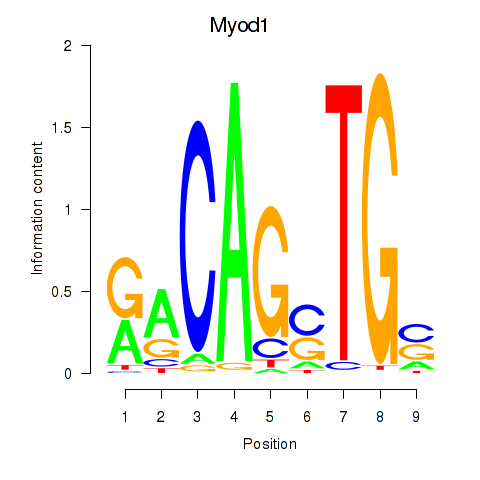
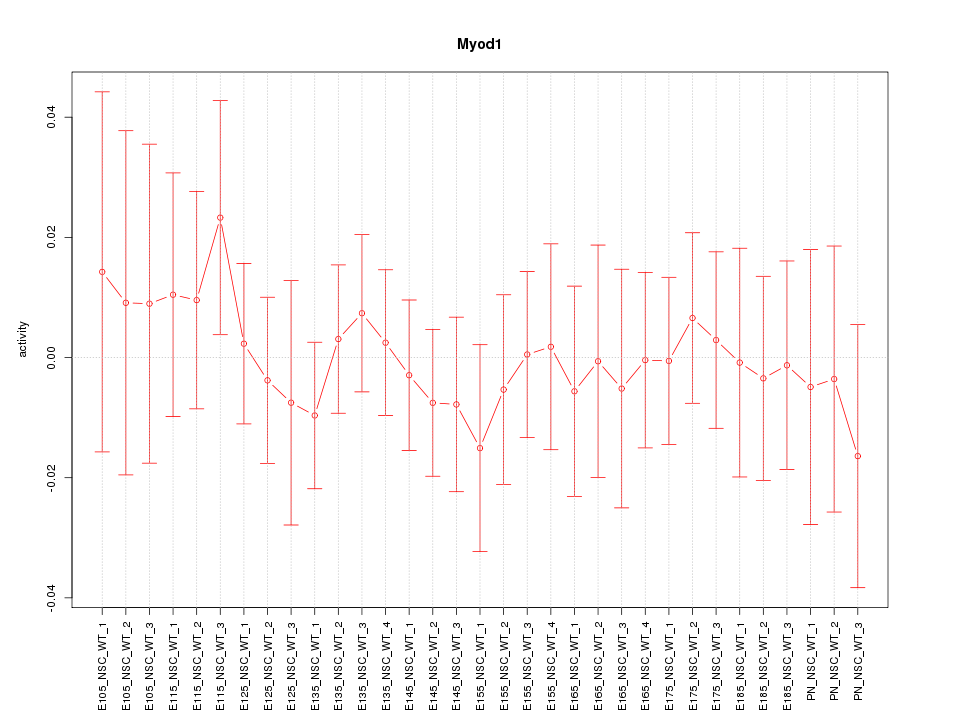
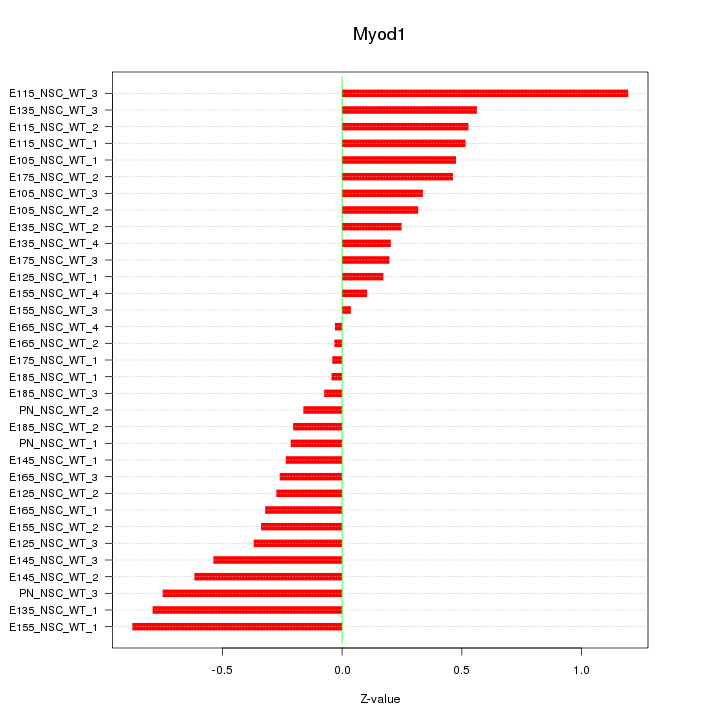
| 1.1 |
3.2 |
GO:0072180 |
mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 0.6 |
1.7 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.5 |
1.4 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.4 |
1.2 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.3 |
4.5 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.3 |
1.4 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.3 |
1.0 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 0.3 |
0.9 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.3 |
0.9 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.3 |
0.9 |
GO:0002159 |
desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) maintenance of organ identity(GO:0048496) |
| 0.3 |
0.6 |
GO:0036023 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 |
1.0 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 |
0.7 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.2 |
0.7 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.2 |
3.0 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 |
0.9 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.2 |
0.9 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.2 |
0.6 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.2 |
3.7 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.2 |
0.5 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 |
1.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 |
0.9 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
0.4 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 |
0.4 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 |
0.8 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.4 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
0.4 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.1 |
1.1 |
GO:0021612 |
facial nerve structural organization(GO:0021612) |
| 0.1 |
0.6 |
GO:0010387 |
COP9 signalosome assembly(GO:0010387) |
| 0.1 |
1.1 |
GO:0021678 |
third ventricle development(GO:0021678) |
| 0.1 |
0.2 |
GO:0072053 |
renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 |
0.4 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.1 |
0.3 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 |
0.7 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
1.3 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
0.3 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.1 |
0.1 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 |
0.3 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.6 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 |
0.8 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
0.4 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 |
0.4 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 |
1.1 |
GO:0014841 |
skeletal muscle satellite cell proliferation(GO:0014841) |
| 0.1 |
0.7 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 |
0.4 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.1 |
0.1 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.1 |
0.7 |
GO:0014866 |
skeletal myofibril assembly(GO:0014866) |
| 0.1 |
0.6 |
GO:0034115 |
negative regulation of heterotypic cell-cell adhesion(GO:0034115) cell-cell adhesion involved in gastrulation(GO:0070586) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 |
0.6 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.1 |
0.3 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.1 |
0.5 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.1 |
0.4 |
GO:0048630 |
skeletal muscle tissue growth(GO:0048630) |
| 0.1 |
0.3 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 |
0.9 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.4 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.3 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 |
0.5 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 |
0.3 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
0.7 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.5 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.1 |
0.2 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 |
0.5 |
GO:0045216 |
cell-cell junction organization(GO:0045216) |
| 0.1 |
0.9 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
0.4 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 |
0.7 |
GO:0046643 |
regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 |
0.4 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
0.5 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.7 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
1.1 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 |
0.8 |
GO:0060033 |
anatomical structure regression(GO:0060033) |
| 0.1 |
0.6 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.1 |
2.0 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 |
0.3 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.4 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 |
0.6 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 |
0.2 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 |
0.3 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.1 |
0.3 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 |
0.8 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.1 |
0.6 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.1 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.1 |
0.3 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 |
0.2 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 |
0.3 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 |
0.1 |
GO:0044849 |
estrous cycle(GO:0044849) cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 |
0.2 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
1.0 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.5 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 |
0.1 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.0 |
0.2 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.5 |
GO:0006670 |
sphingosine metabolic process(GO:0006670) |
| 0.0 |
0.9 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.0 |
0.2 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
1.0 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.2 |
GO:2000258 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
1.3 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.7 |
GO:0042104 |
positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 |
0.1 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 |
0.1 |
GO:0046596 |
regulation of viral entry into host cell(GO:0046596) |
| 0.0 |
0.1 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 |
0.7 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.6 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
1.2 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.1 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.7 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.5 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
0.2 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 |
0.2 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.5 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.1 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 |
0.4 |
GO:1901620 |
regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 |
0.6 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.2 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 |
0.6 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.2 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.7 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.0 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 |
0.2 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.1 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.7 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.0 |
0.4 |
GO:0010666 |
positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 |
0.1 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.0 |
0.2 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.5 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
1.4 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.1 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.0 |
0.2 |
GO:0021924 |
cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 |
0.1 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 |
0.2 |
GO:1902895 |
positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 |
0.1 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.1 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.3 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.6 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 |
0.5 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.0 |
0.3 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.1 |
GO:0046016 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 |
0.4 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.2 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.1 |
GO:0043619 |
regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 |
0.3 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
1.2 |
GO:2001237 |
negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 |
0.1 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 |
0.3 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.4 |
GO:0060135 |
maternal process involved in female pregnancy(GO:0060135) |
| 0.0 |
0.1 |
GO:0002755 |
MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 |
0.2 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 |
0.2 |
GO:0010667 |
negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 |
0.2 |
GO:0015012 |
heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 |
0.1 |
GO:0030889 |
negative regulation of B cell proliferation(GO:0030889) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.2 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.3 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.0 |
0.0 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 |
0.1 |
GO:0045634 |
regulation of melanocyte differentiation(GO:0045634) |
| 0.0 |
0.1 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) |
| 0.0 |
0.2 |
GO:0033006 |
regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 |
0.2 |
GO:0010831 |
positive regulation of myotube differentiation(GO:0010831) |
| 0.0 |
0.1 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.0 |
GO:2000370 |
positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 |
0.2 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.4 |
GO:0006801 |
superoxide metabolic process(GO:0006801) |
| 0.0 |
0.4 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.1 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.0 |
0.1 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.0 |
0.1 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.2 |
GO:0003016 |
respiratory system process(GO:0003016) |