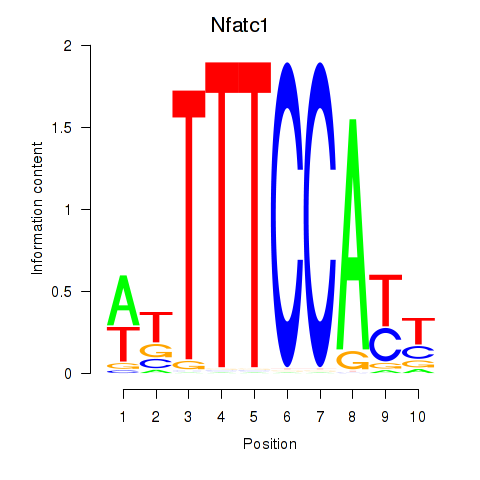
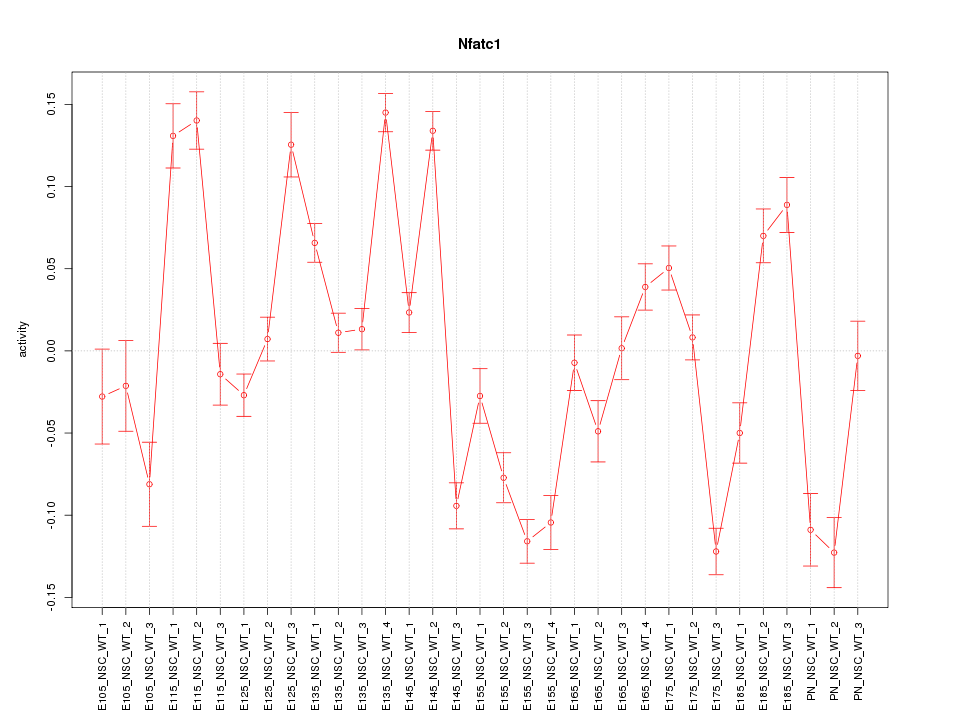
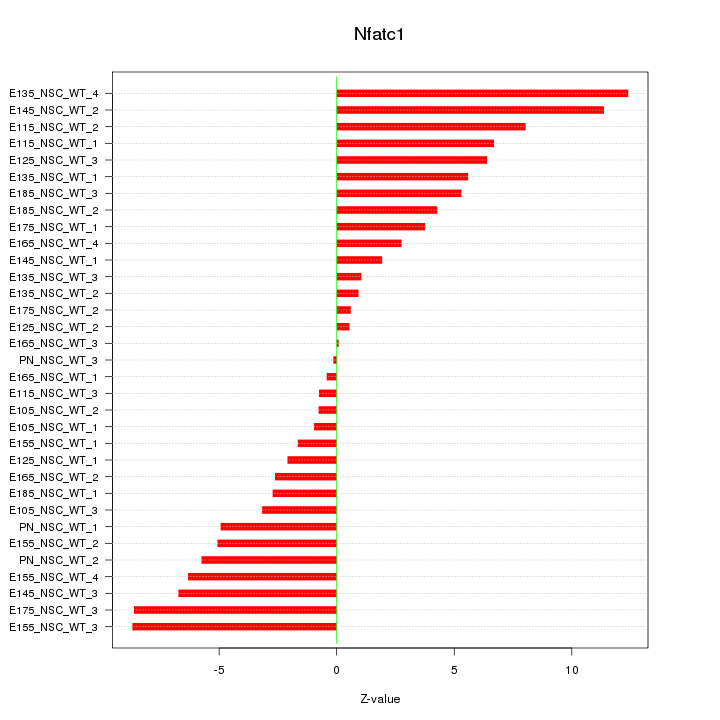
| 2.4 |
9.4 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 2.2 |
15.5 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 2.1 |
6.3 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.9 |
11.1 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 1.8 |
3.6 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.4 |
4.1 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 1.3 |
2.7 |
GO:0060126 |
somatotropin secreting cell differentiation(GO:0060126) |
| 1.2 |
5.0 |
GO:0032439 |
endosome localization(GO:0032439) |
| 1.2 |
3.7 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 1.2 |
4.9 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 1.0 |
2.9 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.9 |
1.8 |
GO:0001787 |
natural killer cell proliferation(GO:0001787) |
| 0.8 |
6.4 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.7 |
7.9 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.6 |
1.7 |
GO:0072139 |
glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.6 |
2.2 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) |
| 0.5 |
3.6 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.5 |
1.0 |
GO:0072034 |
renal vesicle induction(GO:0072034) |
| 0.5 |
0.9 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) |
| 0.4 |
7.7 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.4 |
1.9 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.4 |
2.2 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 |
2.5 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.4 |
0.7 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.3 |
1.0 |
GO:0090135 |
actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) |
| 0.3 |
10.9 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 |
1.5 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.3 |
2.6 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.3 |
4.7 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.3 |
1.3 |
GO:0061181 |
regulation of chondrocyte development(GO:0061181) |
| 0.3 |
1.0 |
GO:0065001 |
negative regulation of histone H3-K36 methylation(GO:0000415) specification of axis polarity(GO:0065001) |
| 0.2 |
0.7 |
GO:0010911 |
regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.2 |
1.1 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.2 |
0.9 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.2 |
2.2 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.2 |
1.9 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.2 |
0.6 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 |
1.1 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.2 |
2.0 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.2 |
0.6 |
GO:1903416 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of calcidiol 1-monooxygenase activity(GO:0060558) response to glycoside(GO:1903416) |
| 0.1 |
0.9 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
1.3 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.1 |
0.8 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.1 |
3.3 |
GO:0048730 |
epidermis morphogenesis(GO:0048730) |
| 0.1 |
1.3 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.1 |
0.9 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
0.6 |
GO:0052695 |
cellular glucuronidation(GO:0052695) |
| 0.1 |
2.0 |
GO:0090103 |
cochlea morphogenesis(GO:0090103) |
| 0.1 |
1.0 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
1.4 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 |
0.5 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 |
0.9 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 |
2.8 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.1 |
0.8 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.1 |
0.6 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 |
0.4 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
2.5 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 |
3.1 |
GO:0042491 |
auditory receptor cell differentiation(GO:0042491) |
| 0.1 |
1.2 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.1 |
1.1 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
0.4 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 |
0.3 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 |
0.3 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 |
1.2 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 |
1.9 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 |
1.1 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
5.5 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.8 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
646.2 |
GO:0008150 |
biological_process(GO:0008150) |