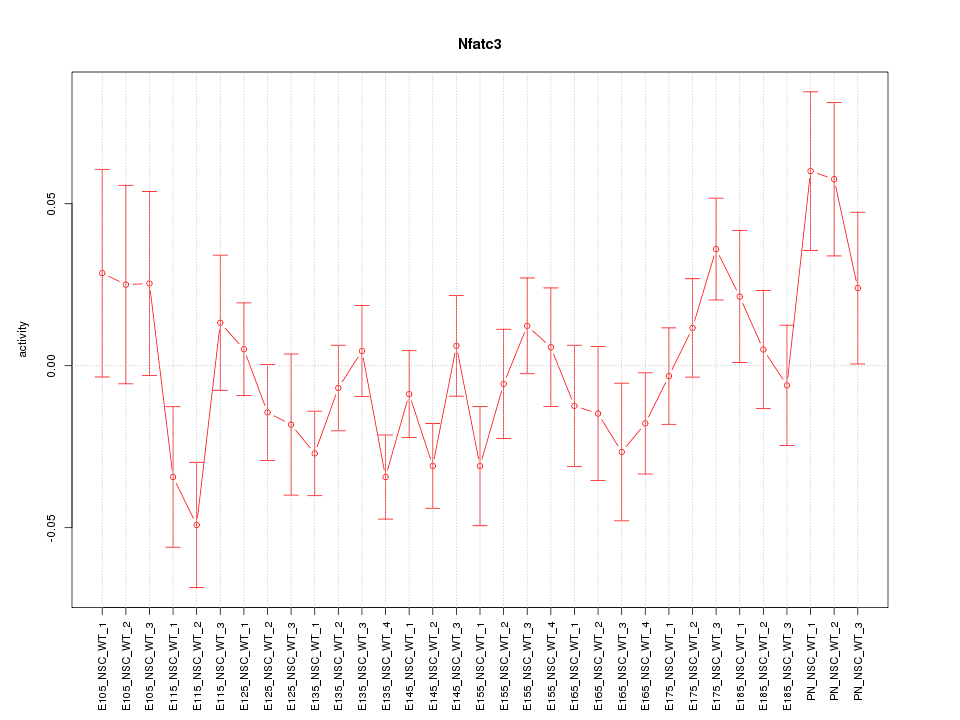
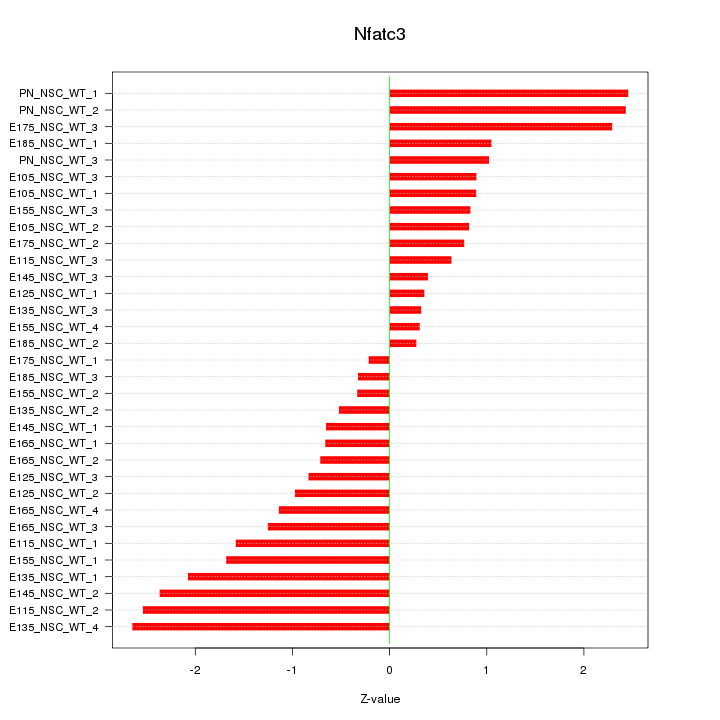
Motif ID: Nfatc3
Z-value: 1.337
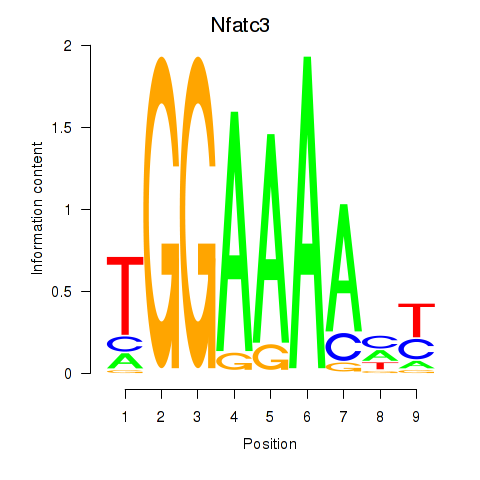
Transcription factors associated with Nfatc3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfatc3 | ENSMUSG00000031902.9 | Nfatc3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc3 | mm10_v2_chr8_+_106059562_106059623 | 0.32 | 7.1e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.3 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 1.9 | 5.6 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 1.8 | 5.3 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 1.2 | 7.0 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.9 | 2.8 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.9 | 1.8 | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.9 | 2.6 | GO:1903903 | striated muscle atrophy(GO:0014891) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of establishment of T cell polarity(GO:1903903) |
| 0.8 | 3.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.7 | 2.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.7 | 2.2 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.7 | 7.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.7 | 3.5 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.7 | 4.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.6 | 4.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.5 | 9.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.5 | 5.9 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.5 | 2.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.5 | 2.8 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.4 | 1.3 | GO:0001803 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) serotonin secretion by platelet(GO:0002554) |
| 0.4 | 1.1 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.4 | 1.1 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 1.9 | GO:1900085 | negative regulation of pinocytosis(GO:0048550) negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.4 | 1.1 | GO:0021557 | oculomotor nerve development(GO:0021557) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.4 | 2.5 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.3 | 1.0 | GO:0046671 | regulation of cellular pH reduction(GO:0032847) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.3 | 2.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 2.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 1.9 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 1.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 2.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.2 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 0.9 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 0.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.2 | 1.3 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.2 | 1.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 1.5 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 1.1 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 | 1.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 | 2.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 0.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 1.0 | GO:2001170 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 0.8 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 4.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 0.7 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 0.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.2 | 1.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.5 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 2.5 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 1.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 1.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 2.7 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 1.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 2.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 2.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.6 | GO:0007135 | meiosis II(GO:0007135) regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.1 | 0.2 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 2.4 | GO:0051894 | integrin activation(GO:0033622) positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.4 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.1 | 1.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.2 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 0.6 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 1.0 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 2.0 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 2.4 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 0.6 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 7.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.4 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 0.2 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.5 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.8 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.4 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.6 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 2.1 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 1.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 2.3 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 2.6 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.4 | GO:0030388 | fructose 6-phosphate metabolic process(GO:0006002) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0031958 | corticosteroid receptor signaling pathway(GO:0031958) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.8 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.5 | GO:0031960 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 | 0.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.6 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 1.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 3.2 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.5 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.8 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.2 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 1.6 | GO:0048754 | branching morphogenesis of an epithelial tube(GO:0048754) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0060047 | heart contraction(GO:0060047) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.5 | 1.5 | GO:0005595 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.4 | 3.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 1.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.4 | 1.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 3.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 9.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.3 | 2.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.0 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 1.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.6 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 5.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 11.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 4.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 4.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 6.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 2.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 1.4 | 7.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.1 | 10.3 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.7 | 2.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.7 | 3.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 7.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.6 | 1.9 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.5 | 4.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.5 | 3.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.5 | 1.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.4 | 1.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.4 | 2.5 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.4 | 1.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.4 | 1.9 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 2.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 2.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 2.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 1.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.7 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.2 | 1.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 1.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.2 | 3.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 2.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.3 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 2.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 2.9 | GO:0050811 | GABA-A receptor activity(GO:0004890) GABA receptor binding(GO:0050811) |
| 0.2 | 1.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 2.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 2.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 2.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.0 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 5.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 4.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0001851 | complement component C3b binding(GO:0001851) proteinase activated receptor binding(GO:0031871) |
| 0.1 | 8.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 0.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.5 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 5.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 3.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0032451 | demethylase activity(GO:0032451) histone demethylase activity(GO:0032452) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.2 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 3.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 3.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 3.0 | GO:0005096 | GTPase activator activity(GO:0005096) |