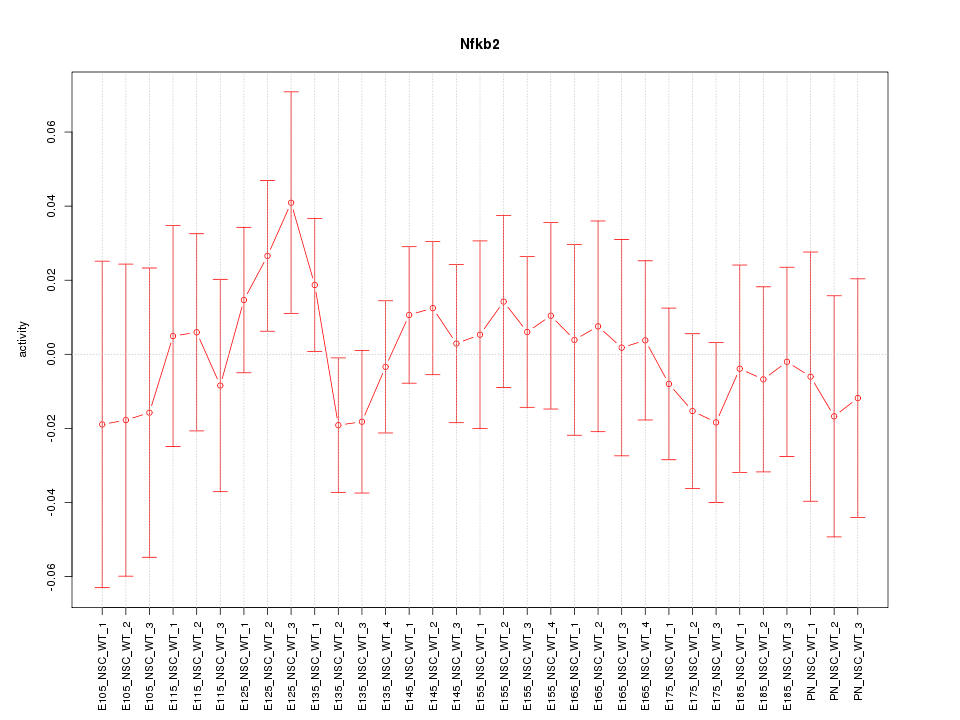
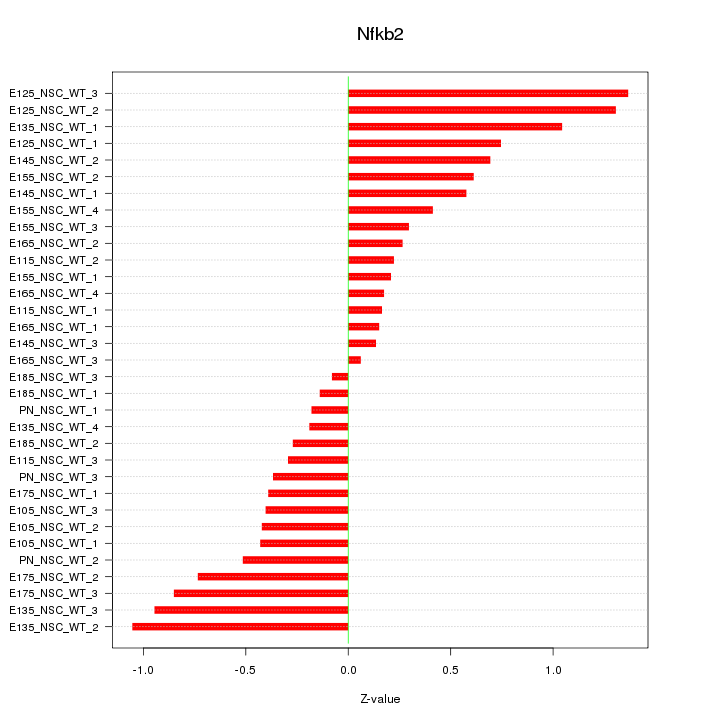
Motif ID: Nfkb2
Z-value: 0.591
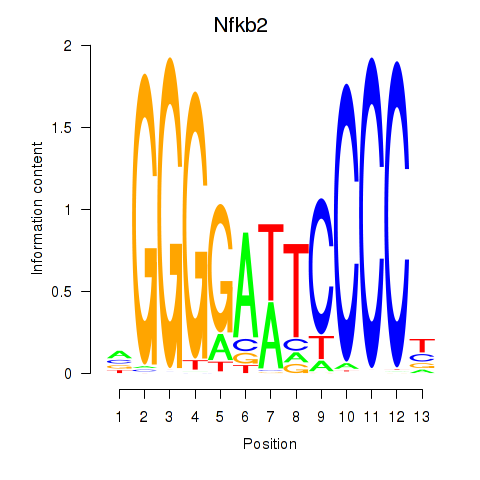
Transcription factors associated with Nfkb2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfkb2 | ENSMUSG00000025225.8 | Nfkb2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb2 | mm10_v2_chr19_+_46304709_46304811 | -0.72 | 2.1e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.3 | 0.9 | GO:0045472 | response to ether(GO:0045472) negative regulation of interleukin-1 beta secretion(GO:0050713) negative regulation of interferon-gamma secretion(GO:1902714) negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.2 | 1.0 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.2 | 1.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) negative regulation of filopodium assembly(GO:0051490) |
| 0.2 | 0.9 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.2 | 0.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 | 0.7 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 1.1 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 | 1.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 2.5 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.8 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.2 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.1 | 0.5 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 2.9 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 1.0 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.5 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.5 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) protein linear polyubiquitination(GO:0097039) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.0 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 0.9 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.3 | 1.2 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.1 | 1.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.8 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 1.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.9 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.2 | GO:0008565 | protein transporter activity(GO:0008565) |