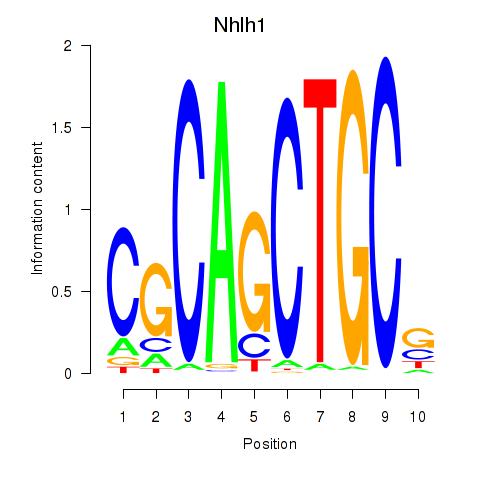
| 3.9 |
15.5 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 2.4 |
7.2 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 2.2 |
2.2 |
GO:0010593 |
negative regulation of lamellipodium assembly(GO:0010593) |
| 1.8 |
5.3 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.7 |
8.3 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 1.7 |
3.3 |
GO:1901204 |
regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 1.5 |
9.0 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 1.5 |
10.4 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.4 |
4.3 |
GO:0046881 |
positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 1.4 |
9.7 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.3 |
3.8 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 1.3 |
3.8 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 1.2 |
3.5 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 1.1 |
1.1 |
GO:0021966 |
corticospinal neuron axon guidance(GO:0021966) |
| 1.1 |
3.3 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.1 |
5.3 |
GO:1902083 |
negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 1.0 |
5.0 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.0 |
3.0 |
GO:1900673 |
olefin metabolic process(GO:1900673) |
| 1.0 |
3.9 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 1.0 |
3.9 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 1.0 |
2.9 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.9 |
7.6 |
GO:0009181 |
purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.9 |
2.8 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.9 |
4.6 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.9 |
2.8 |
GO:0070671 |
response to interleukin-12(GO:0070671) |
| 0.9 |
5.4 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.9 |
3.6 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.9 |
2.7 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.9 |
3.6 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.9 |
4.3 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.9 |
4.3 |
GO:0090467 |
lysine transport(GO:0015819) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) amino acid import into cell(GO:1902837) |
| 0.9 |
2.6 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.9 |
5.1 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.8 |
3.4 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.8 |
0.8 |
GO:0021836 |
chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.8 |
4.1 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.8 |
2.5 |
GO:0070649 |
meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.8 |
1.6 |
GO:0097252 |
oligodendrocyte apoptotic process(GO:0097252) |
| 0.8 |
3.2 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.8 |
3.2 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.8 |
3.9 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.8 |
3.1 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.7 |
5.1 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.7 |
2.9 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.7 |
1.4 |
GO:0046878 |
positive regulation of saliva secretion(GO:0046878) |
| 0.7 |
2.8 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.7 |
5.5 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.7 |
2.1 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.7 |
0.7 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.7 |
2.0 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.7 |
2.0 |
GO:0034334 |
adherens junction maintenance(GO:0034334) |
| 0.6 |
3.9 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.6 |
2.6 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.6 |
6.4 |
GO:0036159 |
inner dynein arm assembly(GO:0036159) |
| 0.6 |
1.9 |
GO:2001170 |
positive regulation of fat cell proliferation(GO:0070346) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.6 |
1.3 |
GO:0006507 |
GPI anchor release(GO:0006507) |
| 0.6 |
1.8 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.6 |
1.8 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.6 |
3.0 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.6 |
1.8 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.6 |
1.7 |
GO:1903726 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.6 |
3.4 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.6 |
2.3 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.6 |
2.8 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.6 |
1.7 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.6 |
1.1 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) |
| 0.5 |
2.7 |
GO:0071072 |
negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.5 |
2.2 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.5 |
1.1 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.5 |
5.3 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.5 |
2.6 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.5 |
3.1 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.5 |
2.6 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.5 |
5.2 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.5 |
2.0 |
GO:2001106 |
regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 |
2.5 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.5 |
2.0 |
GO:0072365 |
regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.5 |
1.4 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.5 |
1.8 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.5 |
1.8 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.5 |
2.7 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.5 |
1.4 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.5 |
1.8 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.4 |
1.8 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.4 |
1.7 |
GO:0060763 |
mammary duct terminal end bud growth(GO:0060763) |
| 0.4 |
1.7 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.4 |
3.0 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.4 |
1.3 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.4 |
0.8 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.4 |
1.7 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.4 |
2.1 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.4 |
8.2 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.4 |
4.9 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.4 |
1.2 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.4 |
2.4 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.4 |
2.8 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.4 |
3.6 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.4 |
1.6 |
GO:0018307 |
enzyme active site formation(GO:0018307) |
| 0.4 |
2.4 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.4 |
2.8 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.4 |
2.7 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.4 |
0.4 |
GO:1904058 |
positive regulation of sensory perception of pain(GO:1904058) |
| 0.4 |
2.3 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.4 |
1.1 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.4 |
2.3 |
GO:0048840 |
otolith development(GO:0048840) |
| 0.4 |
2.3 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.4 |
1.9 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.4 |
1.1 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.4 |
4.5 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) |
| 0.4 |
1.1 |
GO:0039521 |
suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.4 |
3.7 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.4 |
3.3 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.4 |
0.7 |
GO:0014735 |
regulation of muscle atrophy(GO:0014735) |
| 0.4 |
3.7 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.4 |
12.0 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.4 |
0.7 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.4 |
1.8 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.4 |
3.9 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.3 |
2.8 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.3 |
2.1 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.3 |
2.4 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.3 |
2.0 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.3 |
1.0 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.3 |
1.3 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) peptidyl-aspartic acid modification(GO:0018197) |
| 0.3 |
2.0 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 0.3 |
0.7 |
GO:2000974 |
negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.3 |
1.6 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.3 |
3.0 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.3 |
2.3 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 |
1.0 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.3 |
1.0 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.3 |
1.6 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.3 |
1.0 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.3 |
0.6 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.3 |
4.7 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.3 |
0.9 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.3 |
3.4 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.3 |
3.4 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.3 |
4.5 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.3 |
0.9 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.3 |
1.2 |
GO:0050913 |
sensory perception of bitter taste(GO:0050913) |
| 0.3 |
0.6 |
GO:2000344 |
positive regulation of acrosome reaction(GO:2000344) |
| 0.3 |
2.6 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.3 |
2.0 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 |
6.1 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.3 |
1.1 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 |
1.7 |
GO:0010807 |
regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 |
2.7 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.3 |
2.2 |
GO:0060012 |
synaptic transmission, glycinergic(GO:0060012) |
| 0.3 |
3.6 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.3 |
0.8 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.3 |
4.8 |
GO:0010866 |
regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.3 |
1.6 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.3 |
2.7 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.3 |
0.5 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.3 |
0.5 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) ganglion morphogenesis(GO:0061552) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) facioacoustic ganglion development(GO:1903375) dorsal root ganglion morphogenesis(GO:1904835) dorsal root ganglion development(GO:1990791) |
| 0.3 |
0.8 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.3 |
1.3 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 |
2.0 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.3 |
1.3 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 |
1.2 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.2 |
1.5 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) |
| 0.2 |
0.7 |
GO:1903061 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.2 |
1.9 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.2 |
3.8 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.2 |
2.1 |
GO:0023035 |
CD40 signaling pathway(GO:0023035) |
| 0.2 |
0.5 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 |
5.2 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.2 |
0.2 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.2 |
0.7 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 |
3.5 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 |
2.1 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 |
2.5 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.2 |
1.4 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 |
2.1 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.2 |
0.7 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.2 |
1.4 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.2 |
0.7 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 |
0.7 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 |
0.7 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.2 |
0.4 |
GO:0030174 |
regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 |
2.4 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 |
1.6 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.2 |
0.9 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.2 |
0.7 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 |
3.1 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 |
0.4 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 |
1.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.2 |
3.4 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 |
0.8 |
GO:0090283 |
regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.2 |
0.2 |
GO:0014016 |
neuroblast differentiation(GO:0014016) |
| 0.2 |
0.2 |
GO:0006517 |
protein deglycosylation(GO:0006517) |
| 0.2 |
12.0 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.2 |
0.6 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.2 |
1.0 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 |
2.0 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.2 |
4.0 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.2 |
0.8 |
GO:0014819 |
regulation of skeletal muscle contraction(GO:0014819) |
| 0.2 |
1.4 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 |
1.2 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 |
1.0 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.2 |
0.8 |
GO:0007031 |
peroxisome organization(GO:0007031) |
| 0.2 |
1.5 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.2 |
0.7 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 |
3.7 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 |
0.9 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.2 |
0.5 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 |
2.4 |
GO:0006152 |
purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.2 |
1.1 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.2 |
0.7 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.2 |
0.9 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.2 |
1.8 |
GO:0003413 |
chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.2 |
1.2 |
GO:0032411 |
positive regulation of transporter activity(GO:0032411) |
| 0.2 |
1.1 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) |
| 0.2 |
1.1 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.2 |
2.5 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.2 |
3.1 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.2 |
0.9 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 |
1.5 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 |
0.7 |
GO:0045627 |
positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.2 |
1.3 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.2 |
0.8 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.2 |
0.5 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.2 |
2.0 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.2 |
2.5 |
GO:1903779 |
regulation of cardiac conduction(GO:1903779) |
| 0.2 |
13.8 |
GO:0048709 |
oligodendrocyte differentiation(GO:0048709) |
| 0.2 |
0.8 |
GO:0070933 |
histone H4 deacetylation(GO:0070933) |
| 0.2 |
1.8 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.2 |
1.4 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 |
1.1 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.2 |
2.5 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.2 |
0.5 |
GO:0090231 |
regulation of spindle checkpoint(GO:0090231) |
| 0.2 |
0.5 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.2 |
1.4 |
GO:0014898 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.2 |
2.4 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.2 |
0.8 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
1.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
5.5 |
GO:0014037 |
Schwann cell differentiation(GO:0014037) |
| 0.1 |
2.2 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.1 |
1.2 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 |
6.0 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 |
0.3 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.1 |
1.1 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.1 |
0.7 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
1.1 |
GO:0042523 |
positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.1 |
0.7 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
1.2 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.9 |
GO:1904667 |
negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 |
0.9 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.1 |
0.6 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
1.4 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.1 |
4.2 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.1 |
3.8 |
GO:1901381 |
positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 |
1.6 |
GO:0046519 |
sphingoid metabolic process(GO:0046519) |
| 0.1 |
0.7 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 |
0.5 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.1 |
0.4 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.1 |
2.9 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.1 |
1.8 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
0.7 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
1.4 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) |
| 0.1 |
0.3 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
0.9 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
1.6 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.1 |
1.4 |
GO:0044381 |
glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 |
0.3 |
GO:0044791 |
modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 |
2.6 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 |
0.2 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 |
0.3 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 |
0.3 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 |
1.4 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
0.9 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.8 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
1.3 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 |
0.4 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 |
1.7 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.1 |
1.1 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.8 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.3 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 |
3.3 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 |
0.5 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.1 |
2.4 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.1 |
0.5 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
0.5 |
GO:0002681 |
somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 |
0.8 |
GO:0043046 |
DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 |
1.2 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.1 |
0.5 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 |
1.0 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.1 |
0.9 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 |
2.5 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.1 |
0.7 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 |
0.3 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.1 |
0.1 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.1 |
2.1 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.1 |
1.1 |
GO:0090218 |
positive regulation of lipid kinase activity(GO:0090218) |
| 0.1 |
1.9 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 |
2.7 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.1 |
GO:0002586 |
regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 |
0.3 |
GO:0003241 |
growth involved in heart morphogenesis(GO:0003241) |
| 0.1 |
0.9 |
GO:0071801 |
regulation of podosome assembly(GO:0071801) |
| 0.1 |
3.0 |
GO:0051220 |
cytoplasmic sequestering of protein(GO:0051220) |
| 0.1 |
0.6 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 |
1.6 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.1 |
0.9 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.1 |
0.9 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.1 |
2.9 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.1 |
1.1 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
3.5 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.1 |
0.2 |
GO:2000598 |
regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 |
0.2 |
GO:0001547 |
preantral ovarian follicle growth(GO:0001546) antral ovarian follicle growth(GO:0001547) multi-layer follicle stage(GO:0048162) |
| 0.1 |
5.5 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.1 |
0.6 |
GO:0033210 |
leptin-mediated signaling pathway(GO:0033210) |
| 0.1 |
2.0 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.5 |
GO:1904738 |
vascular associated smooth muscle cell migration(GO:1904738) regulation of vascular associated smooth muscle cell migration(GO:1904752) positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.1 |
1.4 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 |
0.3 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 |
0.5 |
GO:0060907 |
positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 |
0.8 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.6 |
GO:1903963 |
arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 |
1.0 |
GO:0009437 |
carnitine metabolic process(GO:0009437) |
| 0.1 |
0.4 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 |
0.7 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.1 |
0.5 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
1.0 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.1 |
0.5 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 |
0.8 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 |
1.1 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
0.3 |
GO:0051956 |
negative regulation of amino acid transport(GO:0051956) |
| 0.1 |
1.8 |
GO:0042058 |
regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 |
0.5 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 |
0.5 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.1 |
1.1 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 |
0.9 |
GO:0022401 |
desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.1 |
0.5 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.1 |
1.0 |
GO:2000058 |
regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 |
0.5 |
GO:0061157 |
mRNA destabilization(GO:0061157) |
| 0.1 |
0.4 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.1 |
0.5 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
0.9 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 |
1.5 |
GO:0071173 |
spindle assembly checkpoint(GO:0071173) |
| 0.1 |
0.4 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.1 |
0.3 |
GO:0043313 |
regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.1 |
0.2 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
0.3 |
GO:0042511 |
positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.1 |
2.0 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.1 |
0.7 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
0.2 |
GO:0008054 |
negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.1 |
0.2 |
GO:0032382 |
positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.1 |
0.9 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.1 |
0.1 |
GO:0046959 |
habituation(GO:0046959) |
| 0.1 |
0.6 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.1 |
0.6 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
2.0 |
GO:0008306 |
associative learning(GO:0008306) |
| 0.0 |
0.1 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 |
0.9 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.6 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.0 |
2.3 |
GO:0035418 |
protein localization to synapse(GO:0035418) |
| 0.0 |
0.7 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 |
1.7 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.0 |
1.1 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.4 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.0 |
0.2 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 |
0.4 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 |
0.8 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.0 |
0.6 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 |
5.8 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
0.7 |
GO:0071357 |
type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 |
0.4 |
GO:0035987 |
endodermal cell differentiation(GO:0035987) |
| 0.0 |
1.5 |
GO:0006998 |
nuclear envelope organization(GO:0006998) |
| 0.0 |
0.7 |
GO:0035249 |
synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 |
0.4 |
GO:0070373 |
negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 |
1.3 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.0 |
0.7 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
0.2 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.6 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
1.0 |
GO:0048278 |
vesicle docking(GO:0048278) |
| 0.0 |
0.3 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
1.2 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.2 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.2 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.5 |
GO:2000641 |
regulation of early endosome to late endosome transport(GO:2000641) |
| 0.0 |
1.2 |
GO:0007613 |
memory(GO:0007613) |
| 0.0 |
0.7 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
0.5 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.5 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 |
1.7 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.1 |
GO:0032466 |
negative regulation of cytokinesis(GO:0032466) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
0.2 |
GO:0006687 |
glycosphingolipid metabolic process(GO:0006687) |
| 0.0 |
0.5 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.3 |
GO:0034088 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 |
0.1 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 |
0.3 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.0 |
0.4 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
0.3 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
1.1 |
GO:0006505 |
GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.9 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.2 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
0.4 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.0 |
0.6 |
GO:0046889 |
positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 |
0.3 |
GO:0042035 |
regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 |
0.1 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.0 |
0.2 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.0 |
0.5 |
GO:0007131 |
reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 |
0.6 |
GO:0061178 |
regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 |
0.6 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.0 |
0.4 |
GO:0005979 |
regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 |
0.8 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.6 |
GO:0090003 |
regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 |
0.2 |
GO:0045806 |
negative regulation of endocytosis(GO:0045806) |
| 0.0 |
0.3 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
0.2 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.9 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.2 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.5 |
GO:1902668 |
negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 |
0.5 |
GO:0021696 |
cerebellar cortex morphogenesis(GO:0021696) |
| 0.0 |
0.4 |
GO:0051647 |
nucleus localization(GO:0051647) |
| 0.0 |
0.4 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.3 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.1 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
3.3 |
GO:0006470 |
protein dephosphorylation(GO:0006470) |
| 0.0 |
0.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.4 |
GO:0002028 |
regulation of sodium ion transport(GO:0002028) |
| 0.0 |
0.3 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.0 |
GO:0048711 |
positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 |
0.5 |
GO:0006040 |
amino sugar metabolic process(GO:0006040) |
| 0.0 |
0.4 |
GO:0051302 |
regulation of cell division(GO:0051302) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.4 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.4 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.0 |
0.2 |
GO:0070232 |
regulation of T cell apoptotic process(GO:0070232) |
| 0.0 |
0.1 |
GO:0044030 |
regulation of DNA methylation(GO:0044030) |