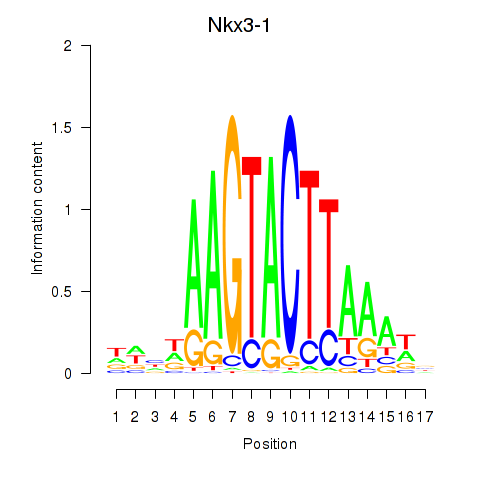
| 1.0 |
3.0 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.8 |
2.3 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.5 |
2.1 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 |
1.5 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.4 |
1.1 |
GO:0002765 |
immune response-inhibiting signal transduction(GO:0002765) |
| 0.3 |
2.8 |
GO:0019240 |
citrulline biosynthetic process(GO:0019240) |
| 0.3 |
0.9 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 0.3 |
0.9 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.3 |
0.8 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.3 |
1.1 |
GO:2000407 |
positive regulation of necrotic cell death(GO:0010940) regulation of T cell extravasation(GO:2000407) |
| 0.3 |
2.0 |
GO:1901550 |
regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.2 |
0.7 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.2 |
1.7 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.2 |
0.7 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 |
0.7 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 |
1.5 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 |
1.0 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.2 |
0.7 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 |
1.2 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 |
1.3 |
GO:2000781 |
positive regulation of double-strand break repair(GO:2000781) |
| 0.1 |
1.9 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.1 |
1.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
1.0 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.5 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.1 |
1.0 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 |
2.0 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.1 |
0.7 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 |
0.6 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
2.9 |
GO:0030224 |
monocyte differentiation(GO:0030224) |
| 0.1 |
1.9 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.1 |
0.7 |
GO:1900116 |
sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 |
1.5 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.1 |
1.0 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 |
0.4 |
GO:0000727 |
double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 |
0.1 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 |
0.9 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 |
0.6 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.1 |
1.0 |
GO:0070633 |
transepithelial transport(GO:0070633) |
| 0.1 |
0.6 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 |
0.9 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 |
1.4 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.1 |
0.2 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
0.2 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.1 |
0.3 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.0 |
1.4 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
1.7 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.2 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.9 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.2 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.0 |
0.3 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.4 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
0.8 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.4 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.0 |
0.3 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.0 |
0.3 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.8 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.7 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 |
0.2 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.0 |
0.1 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.3 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.2 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
0.8 |
GO:0002053 |
positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 |
0.4 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.2 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.3 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) response to cadmium ion(GO:0046686) |
| 0.0 |
0.6 |
GO:0006303 |
double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 |
0.2 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.5 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
3.4 |
GO:0060348 |
bone development(GO:0060348) |
| 0.0 |
0.2 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.1 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.4 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.8 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.0 |
0.3 |
GO:0090382 |
phagosome maturation(GO:0090382) |
| 0.0 |
0.3 |
GO:0043277 |
apoptotic cell clearance(GO:0043277) |
| 0.0 |
0.6 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.8 |
GO:0051289 |
protein homotetramerization(GO:0051289) |
| 0.0 |
0.0 |
GO:0036015 |
response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |