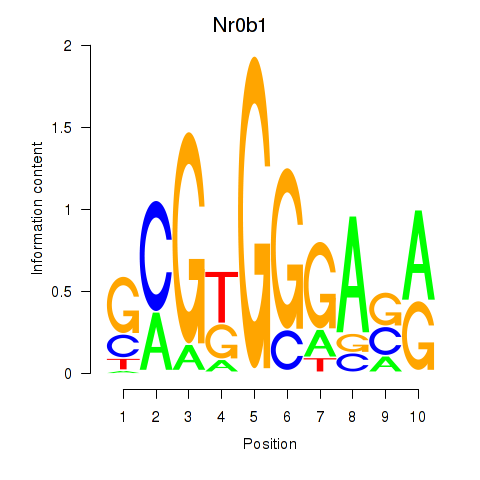
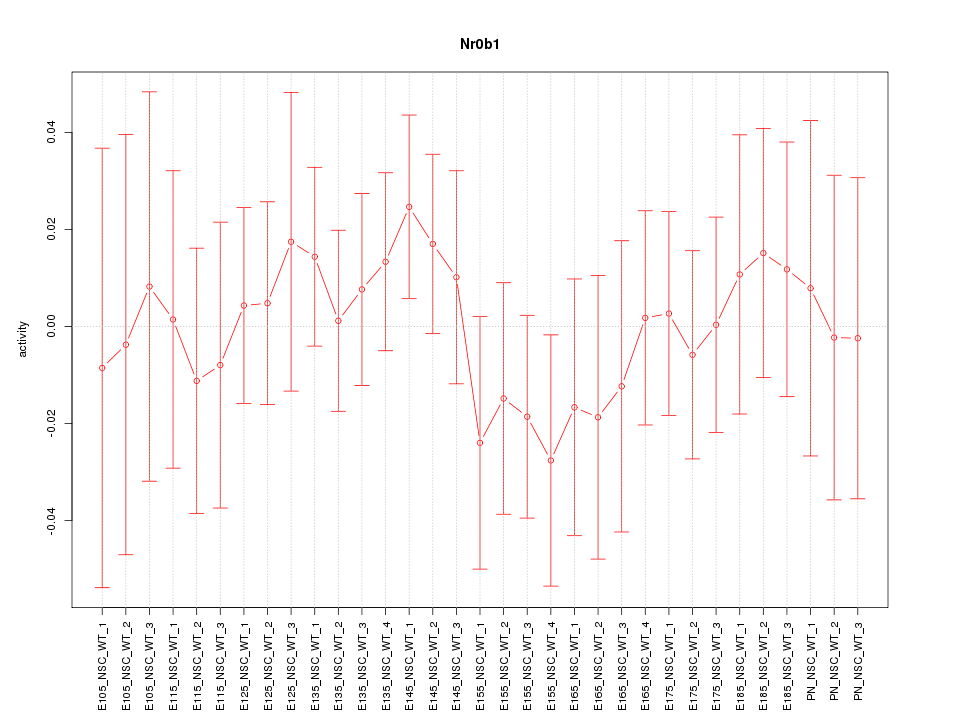
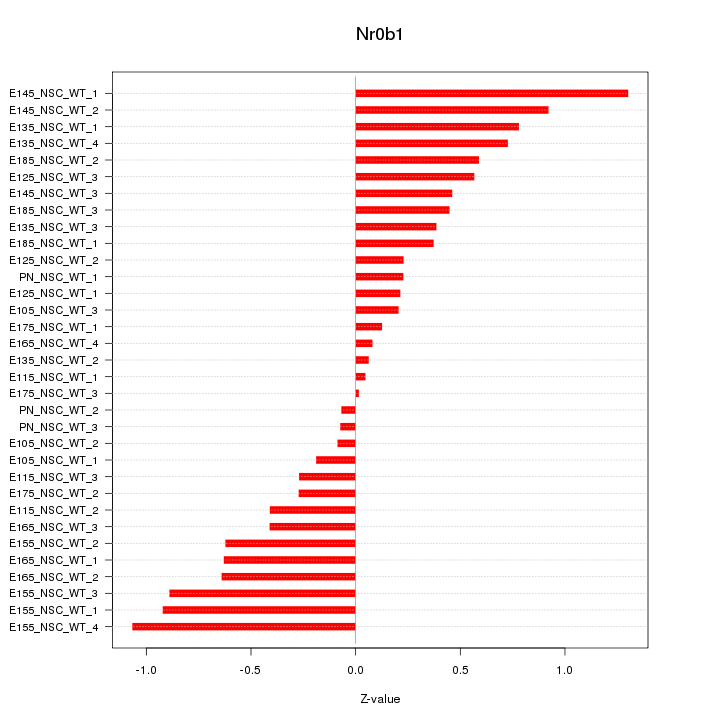
| 0.3 |
1.9 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 |
0.7 |
GO:0072070 |
loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.2 |
2.2 |
GO:0036289 |
peptidyl-serine autophosphorylation(GO:0036289) |
| 0.1 |
1.7 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 |
0.6 |
GO:0007223 |
muscular septum morphogenesis(GO:0003150) Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 |
0.8 |
GO:0051461 |
positive regulation of heat generation(GO:0031652) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
0.4 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.1 |
0.4 |
GO:0051660 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) maintenance of centrosome location(GO:0051661) regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
0.3 |
GO:0035878 |
nail development(GO:0035878) |
| 0.1 |
0.2 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.1 |
0.3 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.1 |
0.3 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 |
0.3 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.1 |
0.6 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.2 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 |
0.2 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 |
0.8 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
0.7 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.1 |
GO:0007354 |
zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 |
0.5 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.1 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 |
0.3 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.2 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 |
0.4 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.3 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.1 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 |
0.7 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.1 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |