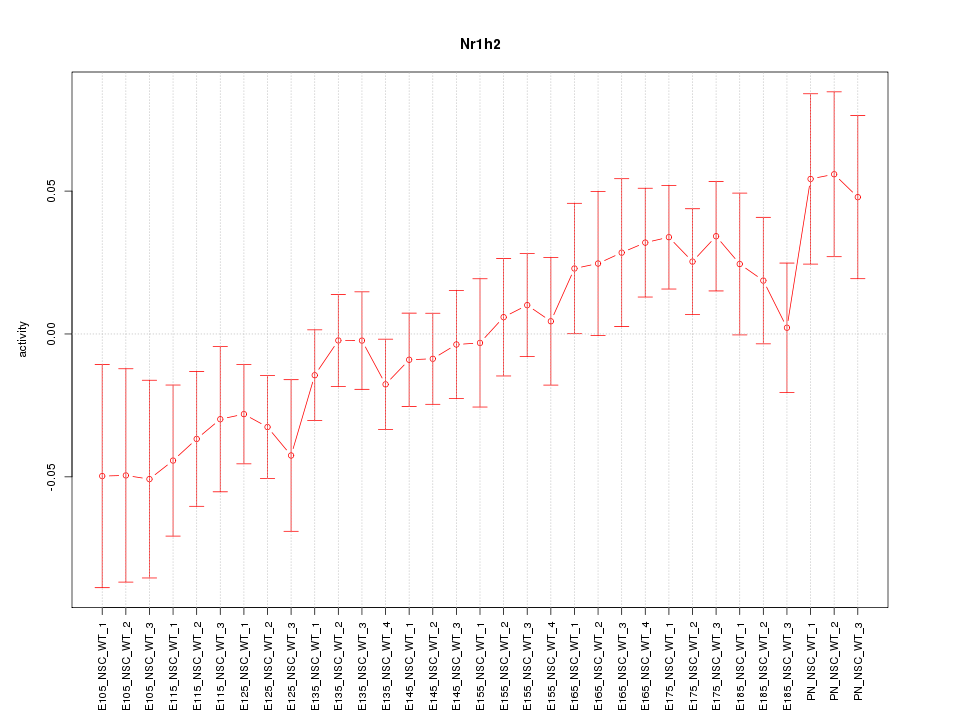
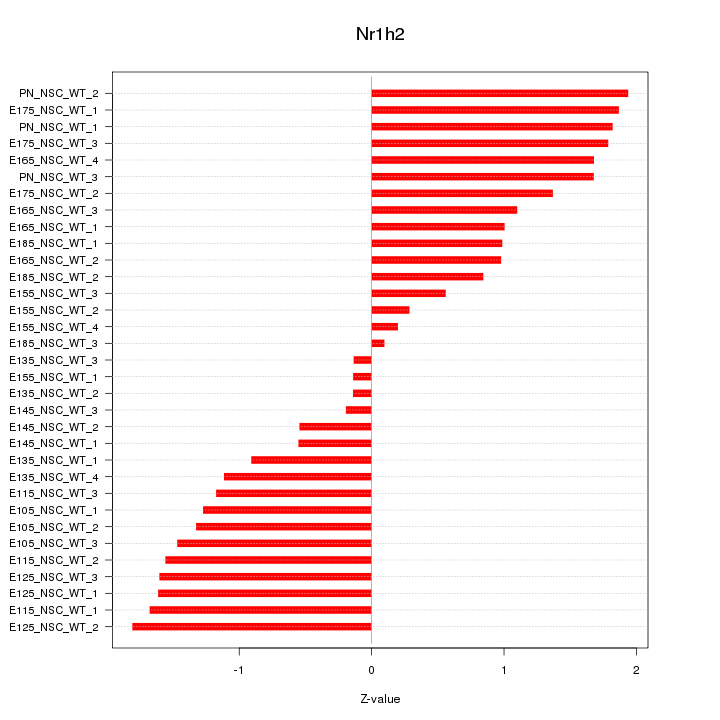
Motif ID: Nr1h2
Z-value: 1.230

Transcription factors associated with Nr1h2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr1h2 | ENSMUSG00000060601.6 | Nr1h2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1h2 | mm10_v2_chr7_-_44553901_44553955 | 0.73 | 1.5e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.5 | GO:0055099 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 2.2 | 6.5 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 1.6 | 4.8 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 1.4 | 7.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 1.3 | 4.0 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 1.3 | 7.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 1.3 | 3.8 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.9 | 2.8 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.8 | 11.2 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.8 | 9.0 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.7 | 9.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.6 | 2.6 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.6 | 1.7 | GO:0014891 | striated muscle atrophy(GO:0014891) |
| 0.6 | 14.7 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.6 | 3.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 1.7 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.4 | 3.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.4 | 1.2 | GO:1902219 | maintenance of blood-brain barrier(GO:0035633) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 | 2.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 1.5 | GO:0010730 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 0.7 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.2 | 1.4 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.2 | 3.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 3.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.2 | 4.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 1.0 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 0.6 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.2 | 2.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 1.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 3.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 1.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 2.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 3.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 1.6 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.4 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 2.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.1 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 1.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 2.5 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.5 | GO:0097006 | regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.0 | 2.1 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 0.1 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.8 | 13.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.6 | 2.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.5 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.5 | 3.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 1.7 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.4 | 2.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 1.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.4 | 2.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 1.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.3 | 1.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 7.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 2.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 2.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 7.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 11.8 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 4.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 6.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 4.4 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 6.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0036126 | sperm flagellum(GO:0036126) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 1.9 | 7.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.4 | 7.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.3 | 4.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 1.1 | 3.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 1.0 | 4.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.9 | 2.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.6 | 6.5 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.6 | 3.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.5 | 2.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 1.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 2.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 1.4 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.3 | 3.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 3.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 1.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 1.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 1.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 0.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.2 | 1.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 1.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.4 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 2.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 1.4 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 3.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 3.2 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 10.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.0 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 1.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 2.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 13.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 2.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 8.2 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 3.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |