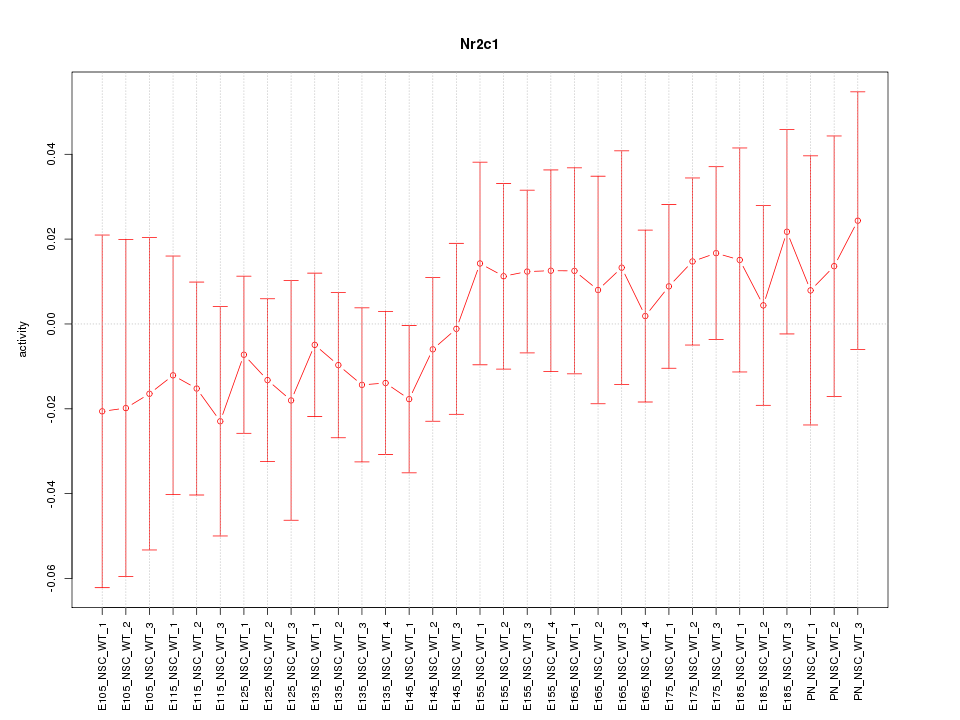
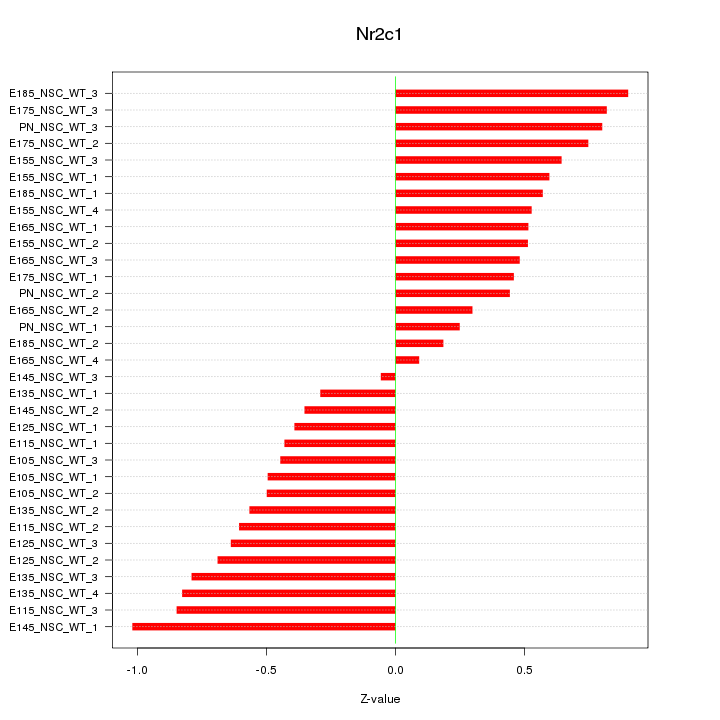
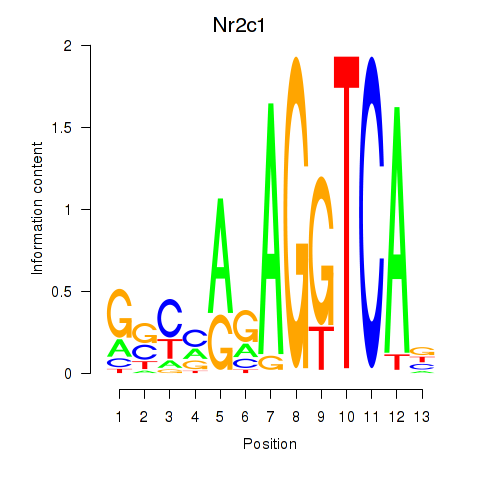
Motif ID: Nr2c1
Z-value: 0.586
Transcription factors associated with Nr2c1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr2c1 | ENSMUSG00000005897.8 | Nr2c1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c1 | mm10_v2_chr10_+_94147982_94148023 | 0.11 | 5.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.5 | 2.6 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.4 | 1.7 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.4 | 3.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 1.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.3 | 2.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 0.7 | GO:1902336 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.2 | 1.4 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.2 | 1.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 1.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 2.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.7 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 2.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.4 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 1.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.4 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.8 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.6 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.0 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.3 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.7 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 4.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.8 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 1.3 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.4 | 2.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 1.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 2.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 4.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 2.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.6 | 2.6 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.4 | 3.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 2.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 2.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.7 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 2.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.9 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 1.1 | GO:0061733 | peptide N-acetyltransferase activity(GO:0034212) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |