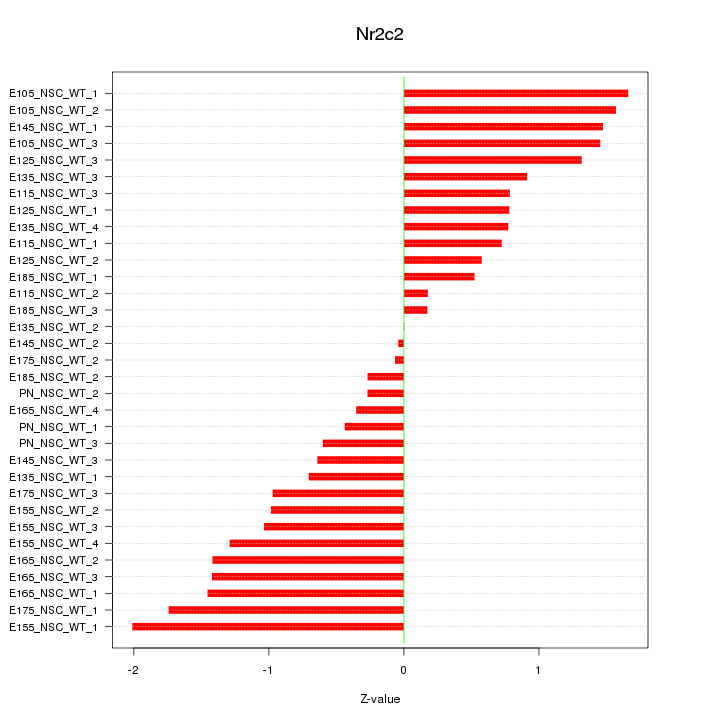
Motif ID: Nr2c2
Z-value: 1.026
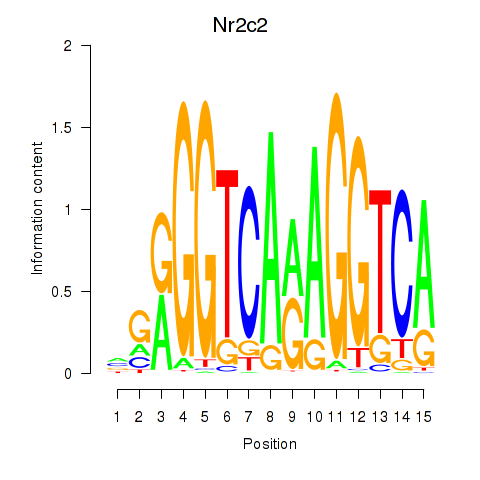
Transcription factors associated with Nr2c2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr2c2 | ENSMUSG00000005893.8 | Nr2c2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c2 | mm10_v2_chr6_+_92091378_92091390 | -0.87 | 3.0e-11 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.5 | 4.6 | GO:0030421 | defecation(GO:0030421) |
| 1.2 | 4.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.0 | 2.9 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.8 | 2.5 | GO:1900211 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) |
| 0.8 | 3.9 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.8 | 4.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.8 | 2.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.7 | 2.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.6 | 6.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 3.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.6 | 1.7 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.6 | 1.7 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.5 | 2.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.5 | 2.7 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.5 | 1.5 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.5 | 3.0 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.5 | 3.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.4 | 4.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.4 | 1.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 2.3 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.4 | 4.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.4 | 1.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.4 | 1.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.4 | 3.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.4 | 2.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.4 | 1.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.4 | 1.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 1.0 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.3 | 0.7 | GO:0071335 | hair follicle cell proliferation(GO:0071335) |
| 0.3 | 2.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.3 | 1.3 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 1.0 | GO:0002842 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.3 | 1.0 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.3 | 1.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 3.6 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.3 | 1.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.3 | 3.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.3 | 0.9 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.3 | 1.5 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.3 | 3.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 0.3 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.3 | 0.8 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.3 | 1.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 0.5 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 1.0 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.3 | 3.6 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.3 | 2.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 0.7 | GO:0072070 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) trachea cartilage morphogenesis(GO:0060535) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.2 | 1.5 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.2 | 5.1 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.2 | 1.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 0.9 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 0.9 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 0.9 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 1.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 2.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.2 | 2.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 0.7 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.2 | 1.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 2.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 1.5 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.2 | 1.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 0.8 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.2 | 0.8 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.2 | 0.8 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 0.6 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.2 | 0.6 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.2 | 1.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 0.5 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 0.9 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 2.0 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.2 | 0.8 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.2 | 2.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 1.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 2.1 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.2 | 1.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 0.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 2.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 1.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 2.4 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 1.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 1.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.4 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 1.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.5 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.3 | GO:0035793 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 1.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.9 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 1.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 1.2 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 2.1 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 | 1.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.0 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.9 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 5.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 1.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 1.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.9 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 0.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.9 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 1.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 2.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.6 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 1.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 2.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.4 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.1 | 1.0 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.4 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.3 | GO:1902626 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 2.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 2.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 2.0 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 1.4 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 2.3 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.1 | 2.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.7 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:0045988 | tachykinin receptor signaling pathway(GO:0007217) negative regulation of striated muscle contraction(GO:0045988) |
| 0.1 | 1.1 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 3.8 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.6 | GO:0015781 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.1 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 2.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 2.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 3.8 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 1.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.0 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 1.0 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 0.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.5 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.1 | 0.8 | GO:0019752 | carboxylic acid metabolic process(GO:0019752) |
| 0.1 | 1.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.2 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 6.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 1.3 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.7 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 1.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.8 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 1.6 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.1 | 1.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.3 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.1 | 0.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.9 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.2 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 1.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 1.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 4.2 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.5 | GO:1904872 | regulation of telomerase RNA localization to Cajal body(GO:1904872) positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 0.0 | 0.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.5 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.1 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.7 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 1.3 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 1.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.4 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.8 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 1.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.3 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0042053 | regulation of dopamine metabolic process(GO:0042053) regulation of catecholamine metabolic process(GO:0042069) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.9 | GO:0097006 | regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 1.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 1.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 3.1 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.3 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.6 | GO:0000077 | DNA damage checkpoint(GO:0000077) DNA integrity checkpoint(GO:0031570) |
| 0.0 | 2.7 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.4 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.5 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.9 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 1.3 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 1.3 | GO:0006997 | nucleus organization(GO:0006997) |
| 0.0 | 0.9 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 2.8 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.6 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 5.1 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.3 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 0.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.6 | 3.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.6 | 2.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.5 | 3.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.5 | 6.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 4.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.4 | 1.3 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.4 | 1.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.4 | 1.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.4 | 2.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.4 | 1.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.4 | 2.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 1.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.3 | 1.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 3.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 1.2 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.2 | 0.7 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 2.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 1.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 1.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 1.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 2.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 1.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.2 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 1.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 3.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 2.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 2.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 0.9 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 1.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.7 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 1.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 5.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.8 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 3.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 2.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 5.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 2.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.4 | GO:0033180 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 2.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 4.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 3.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 3.9 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 7.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.1 | GO:0001891 | phagocytic cup(GO:0001891) podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 15.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 2.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 12.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.2 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 2.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 1.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 4.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 1.9 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.5 | 4.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.2 | 3.6 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 1.1 | 1.1 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 1.1 | 6.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 4.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.8 | 5.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.6 | 2.3 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.5 | 1.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.5 | 0.5 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.5 | 5.2 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.5 | 1.9 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.5 | 1.8 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.4 | 1.2 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.4 | 2.9 | GO:0047631 | adenosine-diphosphatase activity(GO:0043262) ADP-ribose diphosphatase activity(GO:0047631) |
| 0.4 | 1.4 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.4 | 4.3 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.3 | 1.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 3.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 0.9 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.3 | 0.9 | GO:0030362 | protein phosphatase type 4 regulator activity(GO:0030362) |
| 0.3 | 1.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.3 | 1.5 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 2.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 2.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 1.1 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.3 | 2.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 2.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 1.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 0.8 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 2.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 1.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.9 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 2.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 1.5 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.2 | 1.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 2.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 8.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 0.7 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 1.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 3.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 4.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 1.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 0.7 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 0.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 1.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 0.9 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.9 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 4.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 1.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.7 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 1.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.4 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 1.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.9 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.4 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 4.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.8 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.7 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 1.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.1 | 2.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.3 | GO:0016775 | phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 3.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) E-box binding(GO:0070888) |
| 0.1 | 2.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.9 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 2.3 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.1 | 1.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 2.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 4.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 3.4 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 2.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.6 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 4.3 | GO:0030792 | methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 1.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 10.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 2.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 2.3 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 2.6 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 2.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.8 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.0 | 0.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 1.9 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.1 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |