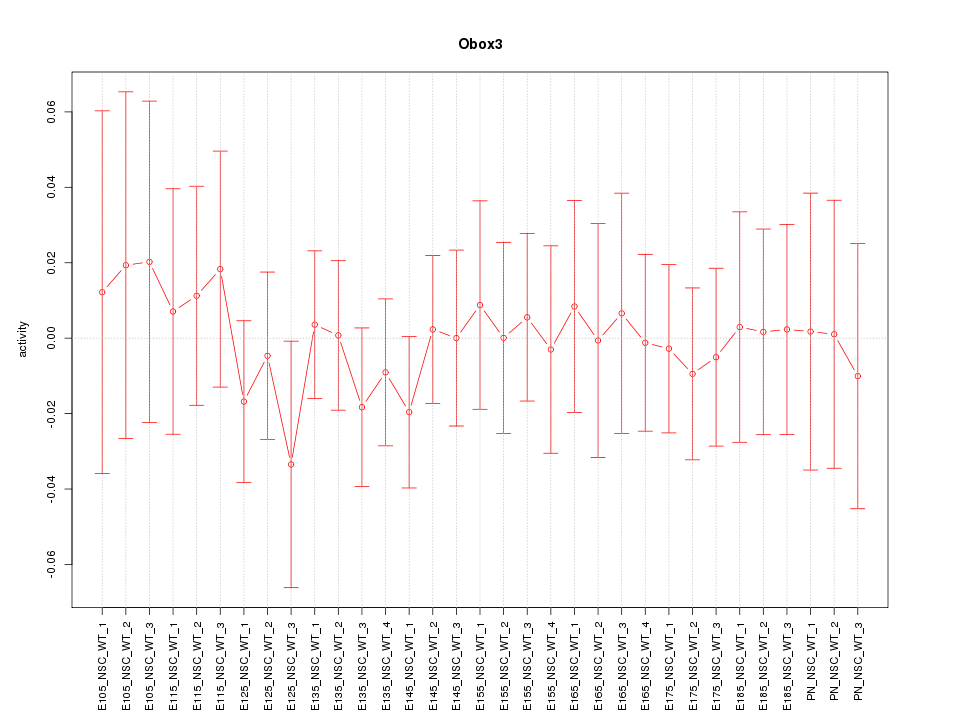
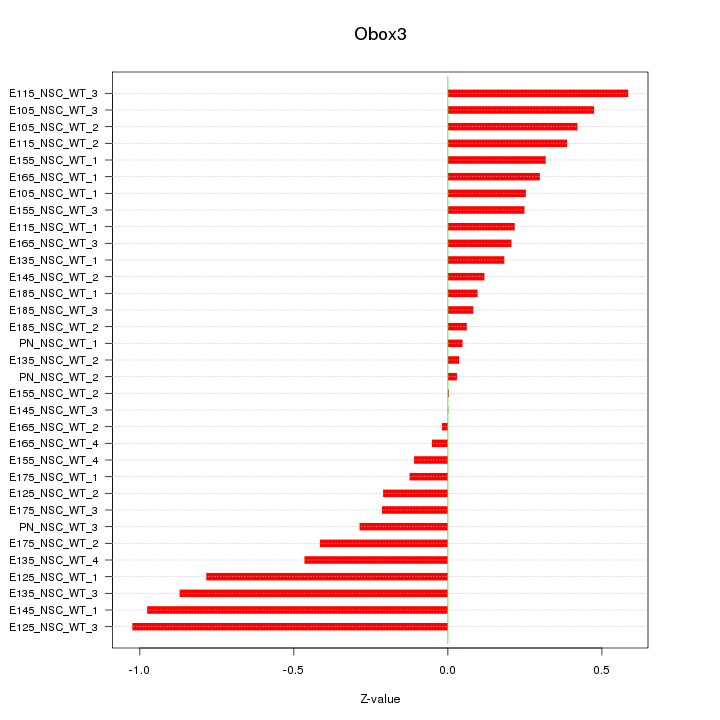
Motif ID: Obox3
Z-value: 0.402
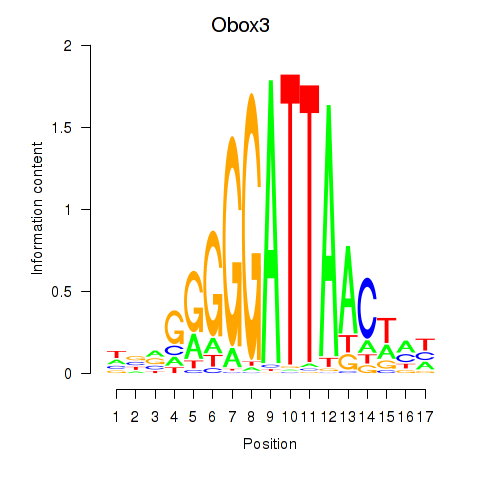
Transcription factors associated with Obox3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Obox3 | ENSMUSG00000066772.6 | Obox3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Obox3 | mm10_v2_chr7_-_15627876_15627876 | -0.16 | 3.9e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.7 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 0.9 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.3 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.5 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) response to folic acid(GO:0051593) |
| 0.1 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.2 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.3 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.4 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.2 | 0.7 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.4 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |