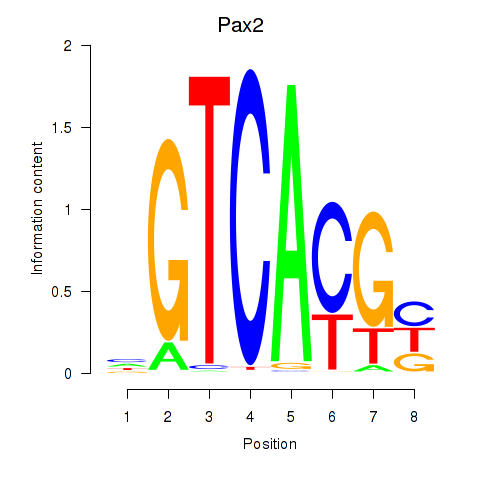
| 1.7 |
5.2 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 1.5 |
4.4 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 1.3 |
3.8 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.9 |
2.8 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.8 |
4.0 |
GO:1900170 |
prolactin signaling pathway(GO:0038161) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.7 |
2.0 |
GO:0033364 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.6 |
2.5 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.6 |
4.2 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.6 |
1.7 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.5 |
2.1 |
GO:2001137 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 0.5 |
2.3 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.4 |
1.2 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.4 |
1.2 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.4 |
2.0 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.4 |
2.0 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.4 |
1.2 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.4 |
1.2 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.3 |
0.3 |
GO:1902513 |
regulation of organelle transport along microtubule(GO:1902513) |
| 0.3 |
4.6 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.2 |
1.2 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.2 |
1.2 |
GO:0051315 |
attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 |
2.1 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.2 |
1.1 |
GO:0019732 |
antifungal humoral response(GO:0019732) |
| 0.2 |
0.4 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.2 |
0.6 |
GO:1904395 |
regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 |
0.6 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.2 |
1.0 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 |
0.6 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.2 |
4.8 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.2 |
1.1 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 |
2.5 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 |
0.7 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.2 |
0.3 |
GO:0071440 |
regulation of histone H3-K14 acetylation(GO:0071440) |
| 0.2 |
0.6 |
GO:0090283 |
regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.2 |
2.8 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 |
0.5 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 |
2.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.2 |
0.8 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
1.5 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 |
0.3 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.1 |
0.5 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 |
1.4 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.9 |
GO:0071376 |
response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 |
0.5 |
GO:0035878 |
nail development(GO:0035878) |
| 0.1 |
0.8 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.1 |
0.5 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
16.6 |
GO:0008643 |
carbohydrate transport(GO:0008643) |
| 0.1 |
0.7 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 |
0.2 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.1 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.1 |
1.3 |
GO:0019243 |
glutathione biosynthetic process(GO:0006750) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 |
0.4 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.9 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.1 |
0.3 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.1 |
1.0 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 |
2.0 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.1 |
0.8 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
1.0 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 |
0.8 |
GO:0003094 |
glomerular filtration(GO:0003094) |
| 0.1 |
0.3 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 |
0.5 |
GO:0090219 |
negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 |
0.2 |
GO:0014012 |
negative regulation of Schwann cell proliferation(GO:0010626) peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 |
0.2 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 |
1.8 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 |
0.5 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
0.3 |
GO:0044829 |
negative regulation by host of viral genome replication(GO:0044828) positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 |
0.3 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.0 |
2.1 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.4 |
GO:0061669 |
spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 |
0.2 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.6 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.2 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 |
0.1 |
GO:0070427 |
nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 |
0.2 |
GO:2000015 |
regulation of determination of dorsal identity(GO:2000015) |
| 0.0 |
0.3 |
GO:1903337 |
positive regulation of vacuolar transport(GO:1903337) |
| 0.0 |
0.5 |
GO:0060055 |
angiogenesis involved in wound healing(GO:0060055) |
| 0.0 |
0.1 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.0 |
0.7 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.8 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.4 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.0 |
0.8 |
GO:0031670 |
cellular response to nutrient(GO:0031670) |
| 0.0 |
0.3 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 |
0.8 |
GO:1903077 |
negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 |
0.2 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.0 |
1.1 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 |
1.7 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.7 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 |
0.9 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.1 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 |
1.0 |
GO:0001656 |
metanephros development(GO:0001656) |
| 0.0 |
0.5 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.0 |
0.9 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.5 |
GO:0045022 |
early endosome to late endosome transport(GO:0045022) |
| 0.0 |
0.6 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
1.1 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.0 |
0.3 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
1.0 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 |
1.3 |
GO:0007286 |
spermatid development(GO:0007286) |
| 0.0 |
0.8 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.4 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.5 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |