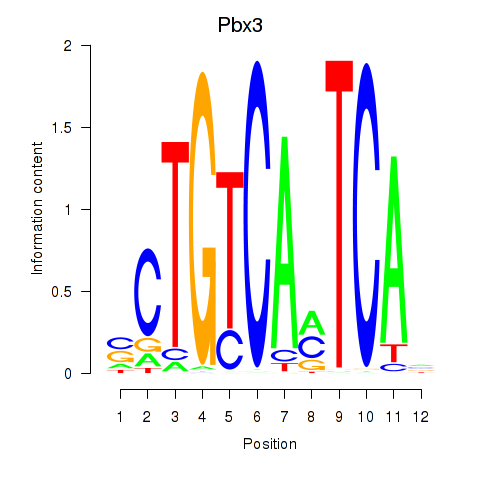
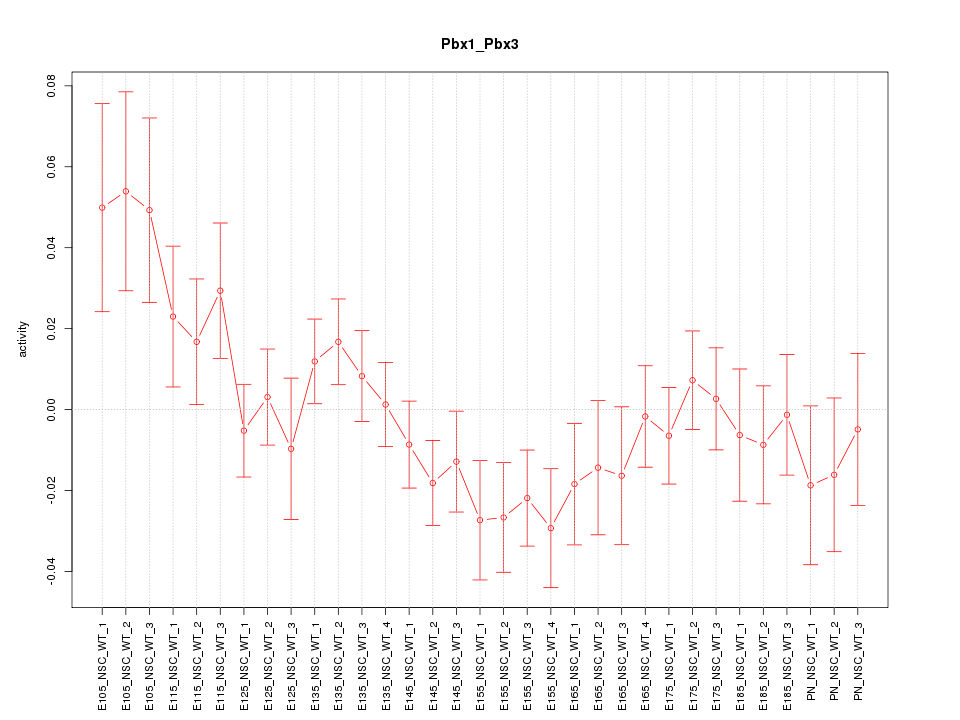
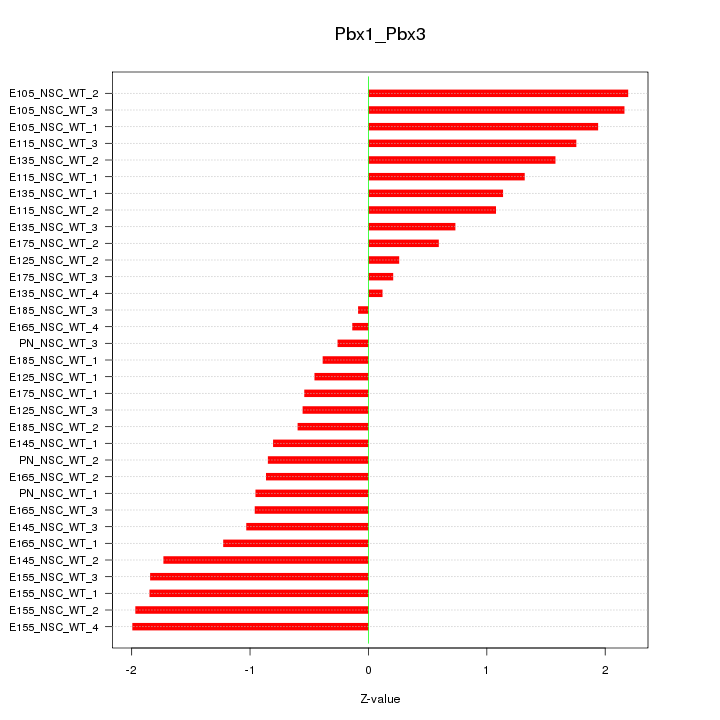
| 3.9 |
11.7 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 3.8 |
15.0 |
GO:0097168 |
condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 3.1 |
9.2 |
GO:0097402 |
neuroblast migration(GO:0097402) |
| 2.1 |
10.7 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 2.1 |
17.1 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 2.1 |
8.3 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.8 |
5.4 |
GO:0060364 |
frontal suture morphogenesis(GO:0060364) |
| 1.5 |
4.6 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 1.1 |
3.4 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 1.0 |
3.1 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 1.0 |
4.1 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.9 |
2.8 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.9 |
2.6 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.8 |
2.4 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.8 |
2.3 |
GO:0045168 |
cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.7 |
3.9 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.6 |
3.2 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.6 |
3.1 |
GO:0048852 |
diencephalon morphogenesis(GO:0048852) |
| 0.5 |
1.6 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.5 |
1.5 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 |
1.4 |
GO:0003223 |
ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.5 |
7.5 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.4 |
3.6 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.4 |
1.8 |
GO:1903898 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.4 |
4.8 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.4 |
2.1 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 |
2.1 |
GO:0006987 |
activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.4 |
1.3 |
GO:0060672 |
negative regulation of keratinocyte differentiation(GO:0045617) epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.4 |
1.2 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.4 |
2.0 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.4 |
6.9 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.4 |
3.8 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 |
1.5 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.4 |
1.9 |
GO:0060066 |
oviduct development(GO:0060066) response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.3 |
1.0 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.3 |
3.4 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.3 |
0.3 |
GO:0061074 |
regulation of neural retina development(GO:0061074) |
| 0.3 |
1.0 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.3 |
1.0 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.3 |
1.0 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.3 |
3.0 |
GO:0030388 |
fructose 6-phosphate metabolic process(GO:0006002) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 |
4.1 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.3 |
0.9 |
GO:0090202 |
transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 |
6.7 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.3 |
1.2 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 |
5.2 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.3 |
0.9 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.3 |
2.0 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.3 |
3.1 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.3 |
1.7 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.3 |
2.6 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 |
0.8 |
GO:0060174 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) limb bud formation(GO:0060174) |
| 0.3 |
2.3 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 |
1.2 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 |
2.1 |
GO:0036123 |
histone H3-K9 dimethylation(GO:0036123) |
| 0.2 |
2.1 |
GO:0075522 |
IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 |
0.7 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 |
2.5 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.2 |
2.4 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 |
0.9 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.2 |
0.9 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 |
0.6 |
GO:0070543 |
response to linoleic acid(GO:0070543) |
| 0.2 |
0.8 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 |
1.0 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.2 |
0.6 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.2 |
1.4 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.2 |
2.0 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 |
1.3 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.2 |
1.9 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.2 |
0.5 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.2 |
0.7 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.2 |
1.3 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 |
0.6 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.2 |
2.5 |
GO:1900745 |
positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 |
2.2 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.2 |
0.5 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 |
0.8 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.2 |
0.3 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.2 |
0.6 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.2 |
2.7 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 |
0.7 |
GO:0060346 |
bone trabecula formation(GO:0060346) |
| 0.1 |
0.6 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 |
1.4 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.1 |
1.0 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.5 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 |
1.7 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 |
2.5 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) |
| 0.1 |
0.4 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 |
0.5 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 |
2.3 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.1 |
0.5 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
2.0 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.1 |
0.3 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 |
6.1 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.1 |
1.3 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
1.5 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 |
0.7 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
1.1 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.1 |
2.7 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
1.1 |
GO:0006573 |
valine metabolic process(GO:0006573) |
| 0.1 |
0.5 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
1.3 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.1 |
1.1 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 |
0.7 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
0.3 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) |
| 0.1 |
3.5 |
GO:0021884 |
forebrain neuron development(GO:0021884) |
| 0.1 |
1.0 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 |
0.4 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 |
1.0 |
GO:0015677 |
copper ion import(GO:0015677) |
| 0.1 |
0.7 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.4 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 |
0.3 |
GO:0097298 |
regulation of nucleus size(GO:0097298) |
| 0.1 |
0.9 |
GO:0043518 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 |
0.9 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 |
0.5 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.1 |
2.2 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.1 |
1.1 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.1 |
1.1 |
GO:2000811 |
negative regulation of anoikis(GO:2000811) |
| 0.1 |
0.7 |
GO:0002098 |
tRNA wobble uridine modification(GO:0002098) |
| 0.1 |
5.0 |
GO:0014066 |
regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 |
0.4 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.1 |
0.4 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 |
0.6 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
2.2 |
GO:1904894 |
positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 |
0.4 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 |
1.4 |
GO:0044342 |
type B pancreatic cell proliferation(GO:0044342) |
| 0.1 |
1.0 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.1 |
0.8 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
1.3 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.4 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 |
0.9 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
2.0 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
2.7 |
GO:0001942 |
hair follicle development(GO:0001942) molting cycle process(GO:0022404) hair cycle process(GO:0022405) skin epidermis development(GO:0098773) |
| 0.0 |
4.7 |
GO:0000910 |
cytokinesis(GO:0000910) |
| 0.0 |
1.2 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.0 |
1.7 |
GO:0006094 |
gluconeogenesis(GO:0006094) |
| 0.0 |
0.2 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 |
0.4 |
GO:0048661 |
positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 |
1.6 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.0 |
7.6 |
GO:0006936 |
muscle contraction(GO:0006936) |
| 0.0 |
0.7 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
1.4 |
GO:0002066 |
columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 |
2.9 |
GO:0030218 |
erythrocyte differentiation(GO:0030218) |
| 0.0 |
2.2 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) |
| 0.0 |
0.4 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 |
0.5 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.2 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
1.1 |
GO:0006801 |
superoxide metabolic process(GO:0006801) |
| 0.0 |
0.1 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.1 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.0 |
0.8 |
GO:0006739 |
NADP metabolic process(GO:0006739) |
| 0.0 |
0.2 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.1 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.0 |
1.6 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.0 |
0.8 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.3 |
GO:0030325 |
adrenal gland development(GO:0030325) |
| 0.0 |
0.6 |
GO:0043687 |
post-translational protein modification(GO:0043687) |
| 0.0 |
0.3 |
GO:0060122 |
inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
0.7 |
GO:0051452 |
intracellular pH reduction(GO:0051452) |
| 0.0 |
0.3 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
0.6 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.1 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.0 |
0.2 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 |
0.5 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
2.0 |
GO:0007219 |
Notch signaling pathway(GO:0007219) |
| 0.0 |
0.1 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 |
0.9 |
GO:0008625 |
extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 |
3.1 |
GO:0001654 |
eye development(GO:0001654) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.0 |
GO:1902177 |
positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 |
0.4 |
GO:0006220 |
pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.0 |
0.1 |
GO:0016126 |
sterol biosynthetic process(GO:0016126) |
| 0.0 |
1.1 |
GO:0006487 |
protein N-linked glycosylation(GO:0006487) |
| 0.0 |
0.8 |
GO:1901343 |
negative regulation of angiogenesis(GO:0016525) negative regulation of vasculature development(GO:1901343) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 |
0.0 |
GO:0070973 |
vesicle coating(GO:0006901) COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 |
4.9 |
GO:0006281 |
DNA repair(GO:0006281) |
| 0.0 |
1.0 |
GO:0090090 |
negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 |
0.1 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.8 |
GO:0001843 |
neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 |
0.5 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.0 |
0.1 |
GO:0001829 |
trophectodermal cell differentiation(GO:0001829) |
| 0.0 |
0.9 |
GO:0006338 |
chromatin remodeling(GO:0006338) |