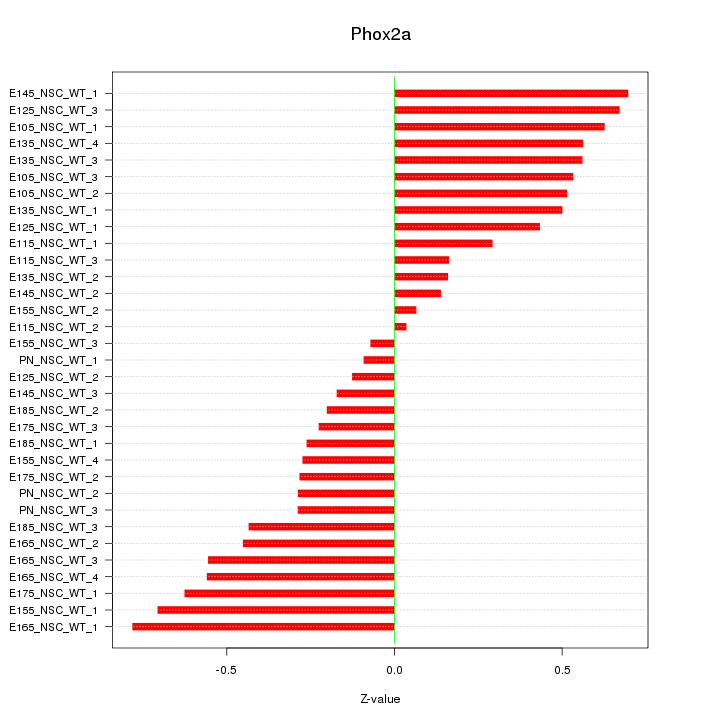
Motif ID: Phox2a
Z-value: 0.432
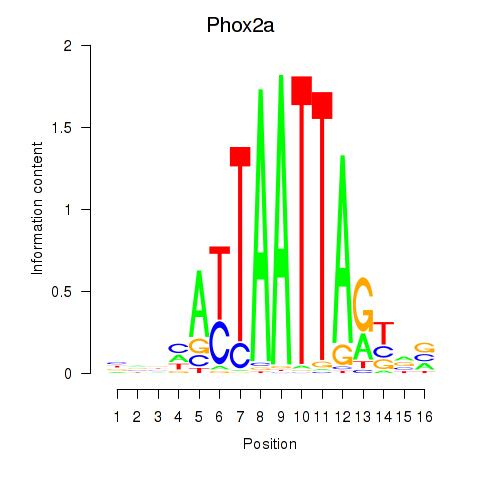
Transcription factors associated with Phox2a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Phox2a | ENSMUSG00000007946.9 | Phox2a |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 0.8 | GO:0060489 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 0.6 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 0.9 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.7 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 1.7 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 2.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 8.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0030689 | preribosome, small subunit precursor(GO:0030688) Noc complex(GO:0030689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 2.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 3.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.7 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |