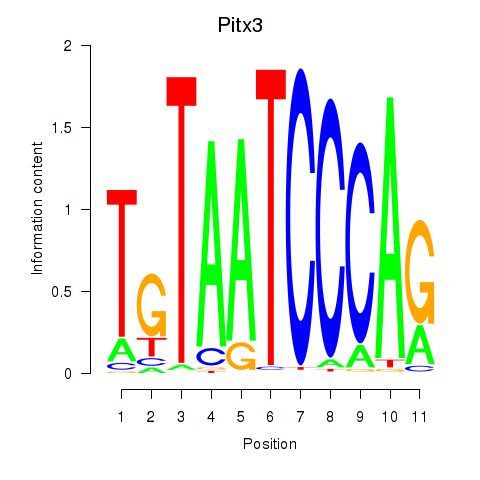
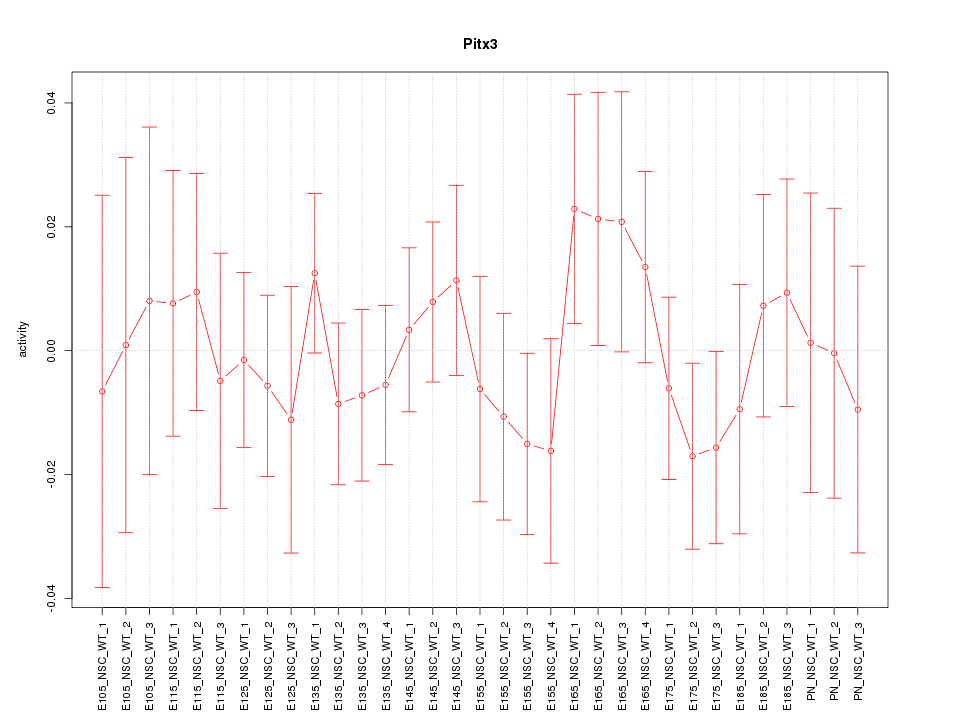
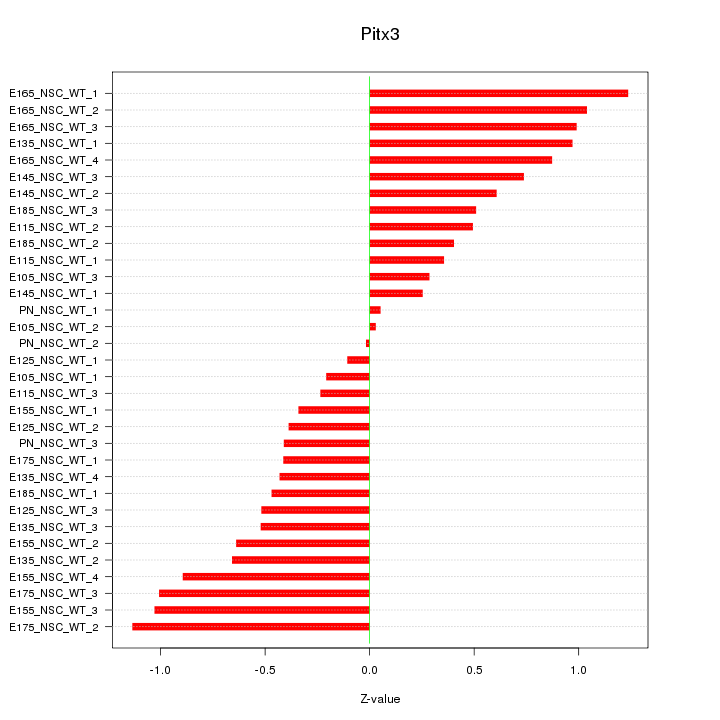
| 0.5 |
2.0 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.5 |
1.4 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.3 |
0.3 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.2 |
1.4 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 |
1.2 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 |
1.1 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.2 |
0.9 |
GO:0044838 |
cell quiescence(GO:0044838) |
| 0.2 |
0.5 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.1 |
0.7 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 |
0.4 |
GO:0002925 |
regulation of dendritic cell cytokine production(GO:0002730) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) detection of peptidoglycan(GO:0032499) negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 |
0.7 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.1 |
0.4 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 |
1.0 |
GO:0002361 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 |
0.5 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 |
0.6 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.1 |
0.4 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 |
0.5 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.1 |
0.3 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.1 |
0.5 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.4 |
GO:1900272 |
negative regulation of chronic inflammatory response(GO:0002677) negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.1 |
0.8 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.1 |
0.3 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 |
0.4 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 |
0.4 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.1 |
0.4 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.1 |
0.3 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 |
0.3 |
GO:0046874 |
quinolinate metabolic process(GO:0046874) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 |
0.2 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 |
0.5 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 |
0.2 |
GO:0000350 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 |
0.2 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
0.2 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 |
0.2 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
0.2 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.1 |
0.3 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 |
0.8 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.1 |
0.2 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 0.1 |
0.3 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.1 |
0.2 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 |
0.3 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.5 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.3 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
0.2 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 |
1.0 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.3 |
GO:0038031 |
non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 |
0.4 |
GO:0014832 |
urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 |
0.2 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 |
0.3 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.3 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 |
0.1 |
GO:0000966 |
RNA 5'-end processing(GO:0000966) |
| 0.0 |
0.1 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.0 |
0.2 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.5 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.1 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 |
0.2 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 |
0.3 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.0 |
0.1 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.3 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.1 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.0 |
0.2 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.0 |
0.7 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.3 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.5 |
GO:2000291 |
regulation of myoblast proliferation(GO:2000291) |
| 0.0 |
0.1 |
GO:1901856 |
negative regulation of cellular respiration(GO:1901856) |
| 0.0 |
0.0 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 |
0.4 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.1 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
0.6 |
GO:0001783 |
B cell apoptotic process(GO:0001783) |
| 0.0 |
2.3 |
GO:0001824 |
blastocyst development(GO:0001824) |
| 0.0 |
0.1 |
GO:0031627 |
telomeric loop formation(GO:0031627) |
| 0.0 |
0.4 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.1 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.3 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 |
0.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.4 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.3 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.5 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.2 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.1 |
GO:0003105 |
negative regulation of glomerular filtration(GO:0003105) |
| 0.0 |
0.3 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.1 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.0 |
0.3 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.1 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.0 |
0.1 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 |
0.1 |
GO:0031282 |
regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 |
0.5 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.0 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
0.3 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.2 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 |
0.1 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 |
0.0 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.0 |
0.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.2 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.0 |
0.1 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.1 |
GO:0033683 |
nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 |
0.1 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.1 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 |
0.1 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.4 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
0.0 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |