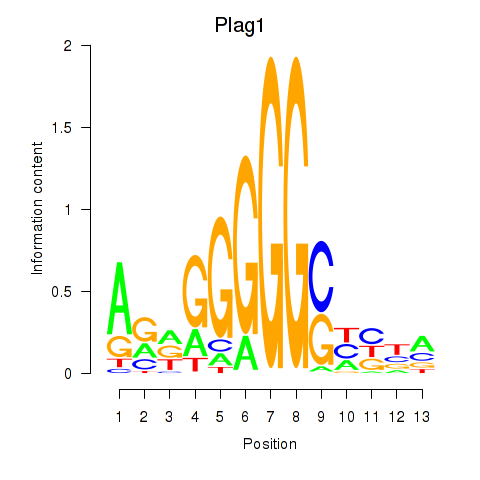
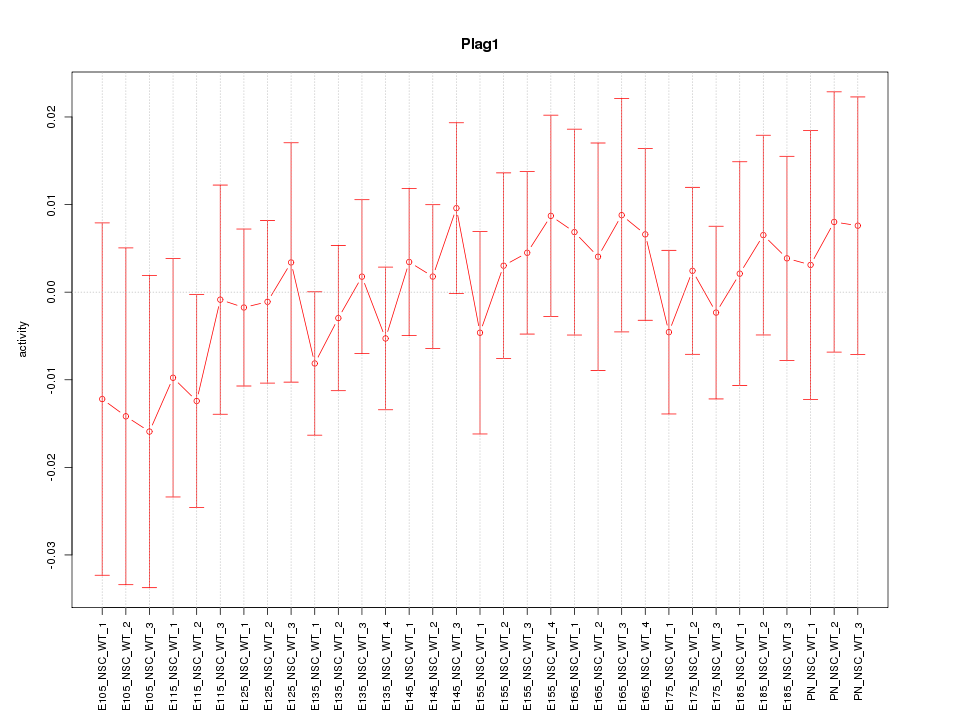
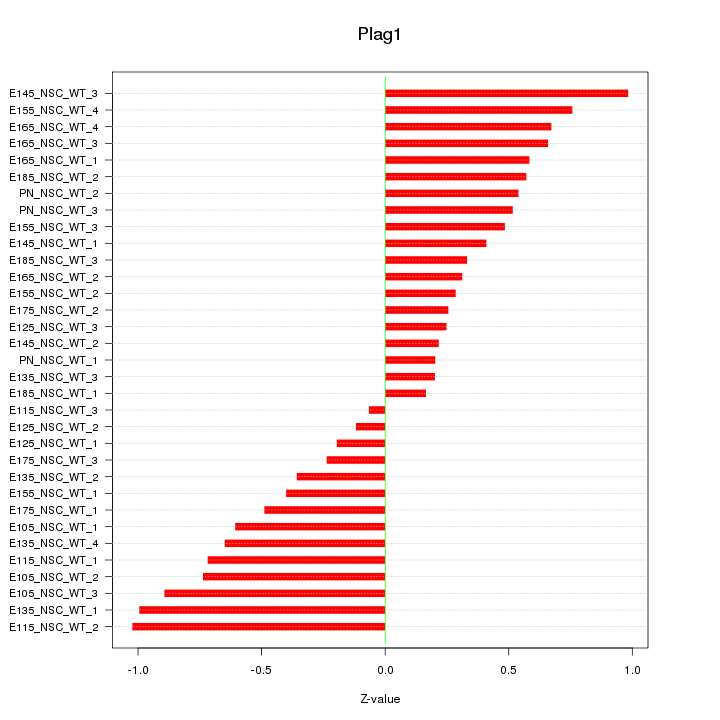
| 2.0 |
5.9 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.7 |
2.8 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.6 |
4.0 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.6 |
2.8 |
GO:0034441 |
plasma lipoprotein particle oxidation(GO:0034441) |
| 0.5 |
1.6 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.4 |
1.3 |
GO:0070671 |
response to interleukin-12(GO:0070671) |
| 0.4 |
1.2 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.4 |
1.5 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.4 |
1.1 |
GO:0051933 |
amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.4 |
1.8 |
GO:0061438 |
renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) |
| 0.4 |
2.5 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.3 |
1.7 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 |
1.0 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.3 |
0.9 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.3 |
0.9 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 |
1.8 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 |
1.1 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.3 |
0.8 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.2 |
1.2 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.2 |
0.7 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.2 |
0.7 |
GO:0060084 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.2 |
0.9 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 |
1.4 |
GO:0060373 |
regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 |
0.6 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.2 |
0.6 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.2 |
0.6 |
GO:1902083 |
negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 |
4.5 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.2 |
2.7 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 |
0.3 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.2 |
0.9 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.2 |
0.5 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.2 |
0.8 |
GO:1902669 |
positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.2 |
0.5 |
GO:0051582 |
positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.2 |
0.5 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 |
1.1 |
GO:1904017 |
response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 |
0.9 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.2 |
2.0 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.2 |
0.8 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 |
0.4 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 |
0.4 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
1.6 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.1 |
0.6 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.1 |
0.7 |
GO:0090494 |
catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.1 |
0.4 |
GO:0015919 |
peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 |
1.5 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 |
1.1 |
GO:2000809 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
1.6 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 |
0.4 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 |
1.0 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 |
0.5 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
0.1 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
0.7 |
GO:1903423 |
positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 |
1.0 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 |
0.6 |
GO:0045188 |
optic nerve morphogenesis(GO:0021631) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.1 |
0.6 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 |
0.6 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.4 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 |
0.6 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
0.3 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.1 |
0.3 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 |
1.3 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.1 |
0.3 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
0.3 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 |
0.3 |
GO:0042939 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 |
0.5 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
0.8 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
0.4 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.6 |
GO:0036258 |
multivesicular body assembly(GO:0036258) |
| 0.1 |
0.3 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 |
0.6 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.1 |
0.9 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.1 |
1.1 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.1 |
0.4 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.1 |
0.3 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.1 |
0.7 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 |
0.3 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
0.4 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 |
0.5 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
1.6 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.1 |
1.2 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 |
0.5 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.1 |
2.0 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
0.4 |
GO:0061428 |
positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 |
4.2 |
GO:0021696 |
cerebellar cortex morphogenesis(GO:0021696) |
| 0.1 |
0.4 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.1 |
0.7 |
GO:0003413 |
chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 |
0.3 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.1 |
2.3 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
0.5 |
GO:0072319 |
clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.1 |
0.4 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.1 |
0.4 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
0.7 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.2 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 |
0.3 |
GO:0015824 |
proline transport(GO:0015824) |
| 0.1 |
0.6 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 |
0.4 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.1 |
0.5 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.1 |
1.7 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.1 |
0.6 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 |
0.9 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.1 |
0.5 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.2 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.1 |
0.1 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
0.1 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 |
0.4 |
GO:1902998 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 |
0.1 |
GO:0061349 |
planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.1 |
0.3 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 |
0.7 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.1 |
0.8 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.1 |
0.3 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 |
0.4 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 |
1.2 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.1 |
0.5 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
0.7 |
GO:0035385 |
Roundabout signaling pathway(GO:0035385) |
| 0.1 |
0.3 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 |
1.1 |
GO:1900746 |
regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 |
0.3 |
GO:0045187 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 |
0.6 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 |
0.4 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.1 |
0.3 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.3 |
GO:1901341 |
activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 |
0.3 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 |
0.1 |
GO:0010643 |
cell communication by chemical coupling(GO:0010643) |
| 0.1 |
0.7 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.1 |
0.8 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.2 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.1 |
0.4 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.1 |
0.4 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.1 |
0.4 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 |
0.2 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.1 |
0.2 |
GO:0071376 |
response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 |
0.2 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.1 |
1.2 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
0.1 |
GO:1902548 |
negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 |
0.3 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.6 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 |
0.4 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
0.2 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.1 |
0.4 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 |
0.5 |
GO:0061577 |
calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 |
0.7 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.1 |
2.5 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 |
0.2 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.3 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.3 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 |
0.7 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.1 |
0.2 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 |
0.6 |
GO:0001956 |
positive regulation of neurotransmitter secretion(GO:0001956) positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 |
0.4 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.0 |
1.7 |
GO:0033344 |
cholesterol efflux(GO:0033344) |
| 0.0 |
0.3 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
1.3 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
0.2 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 |
1.0 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.5 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.2 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.5 |
GO:0050884 |
neuromuscular process controlling posture(GO:0050884) |
| 0.0 |
0.8 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.2 |
GO:0036481 |
intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 |
0.3 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.2 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.4 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.3 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.0 |
1.5 |
GO:1901381 |
positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 |
0.1 |
GO:0046671 |
negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 |
0.2 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.5 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
1.4 |
GO:1902668 |
negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 |
0.2 |
GO:0051012 |
microtubule sliding(GO:0051012) |
| 0.0 |
0.2 |
GO:0032261 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 |
0.2 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 |
0.0 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.0 |
0.6 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.8 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.4 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.1 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 |
0.5 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.0 |
0.5 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.3 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.1 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.2 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.0 |
0.9 |
GO:2001258 |
negative regulation of cation channel activity(GO:2001258) |
| 0.0 |
0.8 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.1 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 |
0.2 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 |
0.9 |
GO:0010043 |
response to zinc ion(GO:0010043) |
| 0.0 |
0.3 |
GO:0033198 |
response to ATP(GO:0033198) |
| 0.0 |
0.1 |
GO:0070445 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 |
0.2 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 |
0.2 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.5 |
GO:0009437 |
carnitine metabolic process(GO:0009437) |
| 0.0 |
0.1 |
GO:0048341 |
paraxial mesoderm formation(GO:0048341) |
| 0.0 |
0.5 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.0 |
0.3 |
GO:0044144 |
regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 |
0.4 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) negative regulation of chromatin binding(GO:0035562) |
| 0.0 |
0.1 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.0 |
0.4 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.4 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
0.6 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.6 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
2.2 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.2 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 |
0.3 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 |
0.1 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.0 |
0.3 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.2 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.0 |
0.3 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.5 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 |
0.1 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 |
0.3 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
0.1 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.0 |
0.3 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 |
0.3 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.1 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.0 |
0.6 |
GO:0051560 |
mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 |
0.8 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.3 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.3 |
GO:0043046 |
DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 |
0.3 |
GO:0035767 |
endothelial cell chemotaxis(GO:0035767) |
| 0.0 |
0.1 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 |
0.4 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.2 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.1 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 |
0.1 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.0 |
0.2 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.4 |
GO:0051383 |
kinetochore organization(GO:0051383) |
| 0.0 |
0.2 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.2 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
3.2 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.6 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.1 |
GO:0033092 |
positive regulation of immature T cell proliferation(GO:0033091) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 |
0.1 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 |
0.3 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 |
0.3 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.1 |
GO:0015911 |
regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) |
| 0.0 |
0.2 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.0 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 |
0.2 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 |
0.2 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.6 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.3 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.2 |
GO:0046785 |
microtubule polymerization(GO:0046785) |
| 0.0 |
0.4 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.4 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 |
0.1 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 |
0.6 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.0 |
0.5 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.0 |
0.2 |
GO:0060004 |
reflex(GO:0060004) |
| 0.0 |
0.5 |
GO:0014068 |
positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 |
0.4 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.4 |
GO:0034243 |
regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 |
0.2 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.0 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.0 |
GO:0060527 |
prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 |
0.6 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 |
0.4 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
1.3 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
0.1 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.3 |
GO:0016242 |
negative regulation of macroautophagy(GO:0016242) |
| 0.0 |
0.4 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
0.9 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
0.3 |
GO:0045453 |
bone resorption(GO:0045453) |
| 0.0 |
0.4 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.3 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.2 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.5 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.2 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.0 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.0 |
0.0 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.0 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 |
0.1 |
GO:0090313 |
regulation of protein targeting to membrane(GO:0090313) positive regulation of protein targeting to membrane(GO:0090314) |
| 0.0 |
0.2 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 |
0.2 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.6 |
GO:0046847 |
filopodium assembly(GO:0046847) |