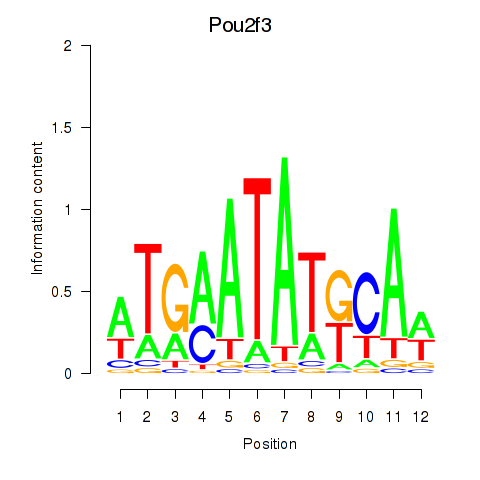
| 1.1 |
3.3 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.8 |
2.4 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.8 |
3.1 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.7 |
4.0 |
GO:0035743 |
CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.5 |
1.6 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.5 |
1.5 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 |
2.4 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.5 |
3.3 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.4 |
1.3 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.4 |
2.5 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.3 |
0.9 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.3 |
0.9 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.3 |
2.1 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 |
1.0 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.2 |
0.9 |
GO:0072015 |
ciliary body morphogenesis(GO:0061073) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 |
0.5 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.1 |
0.9 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 |
0.9 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.1 |
1.5 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.2 |
GO:0003169 |
coronary vein morphogenesis(GO:0003169) |
| 0.1 |
1.6 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
2.4 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.1 |
0.6 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
1.5 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.4 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.1 |
0.5 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
3.0 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
0.3 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.1 |
0.9 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.1 |
0.8 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 |
0.6 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.1 |
0.8 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
0.3 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.1 |
0.2 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 |
0.3 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
0.6 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.6 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 |
0.2 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
0.2 |
GO:2000851 |
positive regulation of cortisol secretion(GO:0051464) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.1 |
0.9 |
GO:0070286 |
axonemal dynein complex assembly(GO:0070286) |
| 0.0 |
0.7 |
GO:0006851 |
mitochondrial calcium ion transport(GO:0006851) |
| 0.0 |
0.2 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 |
0.7 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.3 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.2 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.1 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.0 |
0.7 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
1.1 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.1 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 |
0.3 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.1 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.1 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 |
0.1 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.3 |
GO:2000780 |
negative regulation of double-strand break repair(GO:2000780) |
| 0.0 |
0.4 |
GO:0048246 |
macrophage chemotaxis(GO:0048246) |
| 0.0 |
2.6 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.7 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
1.4 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 |
0.3 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.7 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 |
1.1 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.4 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.1 |
GO:0032382 |
positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.0 |
0.4 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.1 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 |
0.4 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.0 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
1.8 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.3 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |